CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 47
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM47.html |
Ligand specificities
The characterization of the C-terminal triplet fucose-binding module (SpX-1.2.3) of protein toxin SpGH98, originating from the fucose utilization operon in Streptococcus pneumoniae, was the founding member of the CBM47 family [1]. However, the Anguilla anguilla agglutinin (AAA) derived from the European eel, which was characterized earlier [2], is also included in the CBM47 family. Several other members of the CBM47 family, such as MsaFBP32 [3], LLYlec [4], and its mutant LLYlecY62H [5], have been confirmed to possess the F-type lectin fold. The aforementioned proteins are all capable of binding to carbohydrates containing fucose, which could be oligosaccharides (often trisaccharides or smaller) or parts of polysaccharides, such as the AAA, and a portion of CBM47 members have demonstrated binding to glycans containing galactose [2]. Both of these sugars are ubiquitous in glycoproteins and glycolipids on the cell surface, playing crucial roles in cellular bioactivity and functions. Furthermore, the CBM47 family exhibits specific binding to Lewis blood group oligosaccharides, which are characterized by fucosylation modifications. For instance, LLYlec binds to both Lewis y (Ley) antigen (type 1 antigen) and Lewis b (Leb) antigen (type 2 antigen) [4], whereas AAA displays a preference for binding to the Ley antigen [2]. Consequently, the CBM47 family holds potential applications in immune recognition and disease diagnosis. Besides, another CBM47 domain (named WfCBM47) was discovered from the marine bacterium Wenyingzhuangia fucanilytica CZ1127T, which is appended to the GH168 family sequence. The CBM binds to the various sulfated fucans with the backbone composed of 1,3-α-L-fucopyranose residues [6].
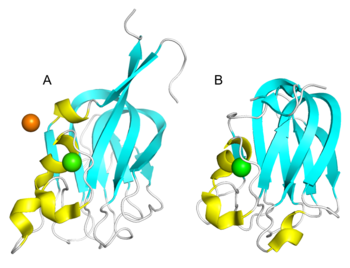
Structural Features
All the fucose-binding proteins, SpX-1 (PDB 2j1r), SpX-3 (PDB 2j22) and AAA (PDB 1k12), have had their tertiary structures determined through X-ray crystallography [1, 2]. The core structures of the CBM47 family exhibit remarkable similarity, adopting an eight-stranded β-sandwich fold, which is comprised of a five-stranded anti-parallel β-sheet on one side and a three-stranded anti-parallel β-sheet on the other [1, 2, 3, 4, 5]. Additionally, CBM47 typically exists as a dimer or trimer under physiological conditions, with Ca2+ playing a pivotal role in maintaining its conformational stability [1, 2, 4]. The CBM47 family recognizes and binds glycans through shallow grooves located within their complementarity-determining regions (CDRs); these variable loops have been shown to exhibit a high degree of sequence and conformational variability among different CBM47s, enabling them to recognize diverse ligands [4]. For instance, LLYlec and SpX-1 reveal smaller differences among their loops, permitting them to bind to a wide array of Lewis blood group oligosaccharides [1, 4]. While AAA features a particularly longer and rigid loop, which may account for its preference to form complexes with type 1 antigens while lacking affinity for type 2 antigens [2]. Additionally, the loop regions of MsaFBP32 are shorter than those of AAA, resulting in fewer steric hindrances, which facilitates easier contact with oligosaccharide molecules and the formation of stable binding, but concurrently may reduce binding specificity [3].
Functionalities
The AAA, discovered in the serum of European eel, and MsaFBP32, isolated from the plasma of the striped bass Morone saxatilis, participate in the recognition of bacterial lipopolysaccharides by the animal innate immune system. For instance, they are capable of recognizing and binding to specific carbohydrate structures on the surfaces of pathogens, such as L-fucose and D-galactose methylether derivatives, serving as vital immune recognition molecules. Furthermore, they can also recognize H and Lea antigens, which are used in determining ABO and Lewis blood group types [2, 3].
Both LLYlec and SpX-1.2.3 are associated with bacterial toxins belonging to the GH98 family. Under physiological conditions, these proteins form aggregates that bind to glycoproteins or glycolipids on the cell membrane. These aggregates, through protein-protein interactions with the toxin, steer the toxin to accumulate at fucose-rich sites on the cell surface, thereby enhancing the pore-forming activity of the toxins and accelerating cell lysis. Based on the binding specificity to fucoidan, the fluorescein isothiocyanate-labeled SpX-1.2.3 triplet was applied for in situ visualization of mouse lung tissue sections [1, 4].
Sulfated fucans, as one of the structural molecules in the sea cucumber body wall, play an important role in the formation of the structure and quality of sea cucumber. A fluorescent probe was constructed by fusing WfCBM47 with a green fluorescent protein, based on which the in situ visualization of sulfated fucan in the sea cucumber (Apostichopus japonicus) body wall was implemented for the first time [6]. It was observed that sulfated fucan existed as a ground substance in the extracellular matrix.
Family Firsts
- First Identified
- SpX-1.2.3, isolated from Streptococcus pneumoniae and associated with the toxin SpGH98, was reported in 2006 and is considered the founding member of the CBM47 family [1]. The F-type lectin AAA, discovered in the serum of European eel and reported in 2002, is also included in the CBM47 family [2].
- First Structural Characterization
- The first structural characterization was on the AAA (PDB 1k12) prior to the founding of the CBM47 family [2].
References
- Boraston AB, Wang D, and Burke RD. (2006). Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006;281(46):35263-71. DOI:10.1074/jbc.M607620200 |
- Bianchet MA, Odom EW, Vasta GR, and Amzel LM. (2002). A novel fucose recognition fold involved in innate immunity. Nat Struct Biol. 2002;9(8):628-34. DOI:10.1038/nsb817 |
- Bianchet MA, Odom EW, Vasta GR, and Amzel LM. (2010). Structure and specificity of a binary tandem domain F-lectin from striped bass (Morone saxatilis). J Mol Biol. 2010;401(2):239-52. DOI:10.1016/j.jmb.2010.06.018 |
- Feil SC, Lawrence S, Mulhern TD, Holien JK, Hotze EM, Farrand S, Tweten RK, and Parker MW. (2012). Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity. Structure. 2012;20(2):248-58. DOI:10.1016/j.str.2011.11.017 |
- Lawrence SL, Feil SC, Holien JK, Kuiper MJ, Doughty L, Dolezal O, Mulhern TD, Tweten RK, and Parker MW. (2012). Manipulating the Lewis antigen specificity of the cholesterol-dependent cytolysin lectinolysin. Front Immunol. 2012;3:330. DOI:10.3389/fimmu.2012.00330 |
- Mei X, Chang Y, Shen J, Zhang Y, Chen G, Liu Y, and Xue C. (2022). Characterization of a sulfated fucan-specific carbohydrate-binding module: A promising tool for investigating sulfated fucans. Carbohydr Polym. 2022;277:118748. DOI:10.1016/j.carbpol.2021.118748 |