CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 63
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM63.html |
Ligand specificities
In nature, CBM63s are found almost exclusively as the C-terminal domain of two-domain proteins named expansins [1]. Expansins are ubiquitous in plants where they function in cell growth and other developmental processes that involve plant cell wall modification [1, 2]. They are also present in diverse microbes, including many bacterial and fungal plant pathogens [3]. Microbes may have acquired expansin genes via horizontal gene transfer(s) from plants in the distant past [4]. In the current CAZypedia database, plant sequences are not included in the CBM63 listings, possibly a consequence of the large sequence divergence between bacterial and plant expansins, which might hinder automated sequence-based methods used to construct CBM families. However, structural analyses (see below) leave little doubt of the homology between plant and bacterial expansins [5], including the C-terminal domain identified as CBM63 in numerous microbial expansins.
The most detailed characterization of CBM63 is based on the C-terminus of the expansin BsEXLX1 produced by the soil bacterium Bacillus subtilis [6] (GenBank: AAB84448.1). It contains ~100 amino acids with a molecular weight of ~11.5 kDa. Recombinant protein (the entire two-domain protein, without histidine tags) binds cellulose (Avicel) with an affinity (KD) of 2.1 μM and binding capacity (Bmax) of 0.3 μmol/g of cellulose [7]. Virtually identical binding parameters were found for the recombinant CBM63 alone, whereas binding of the N-terminal domain was below the detection limit of the depletion isotherm assay [7]. Evidently BsEXLX1 binding to cellulose is solely determined by the CBM63 domain [7].
The two-domain BsEXLX1 protein also binds to whole cell walls from wheat coleoptiles, primarily via the CBM63 domain, with an affinity (KD) of 1.79 μM and the remarkably high binding capacity of 30 μmol/g (0.345 g/g) of cell wall [7]. Hence, whole cell walls have 100-fold greater binding capacity for CBM63 compared to Avicel, on a dry weight basis. This high capacity may be partly the result of differences in cellulose structure, aggregation and accessibility, but more likely the higher capacity results primarily from abundant interactions with the matrix, including electrostatic interactions of this basic protein (pI ~ 9.7) with pectins and other acidic matrix polysaccharides [7]. Site-directed mutagenesis and additional binding experiments [7] show that the high binding capacity of wheat cell walls depends on electrostatic interactions between basic residues (K145, K171, R173, K180, K183, K188) on the ‘back side’ of the CBM63 domain (the ‘front side’ is defined as the surface that binds cellulose; see figure 1) and matrix polysaccharides (predominantly glucuronoarabinoxylan and pectins). Consistent with this conclusion, binding to coleoptile cell wall was reduced 95% in the presence of 10 mM CaCl2, whereas binding to cellulose was reduced only ~40% in CaCl2 (up to 100 mM) [7]. Moreover, wild-type BsEXLX1 binds insoluble arabinoxylan from wheat flour (KD 4.7 μM; Bmax 3.3 μmol/g) [7]. This binding was eliminated when three of the basic residues listed above were changed to Q [7].
The above studies show that CBM63 from BsEXLX1 has separate surfaces that bind cellulose and matrix polysaccharides (e.g. arabinoxylan) through distinct physical interactions. In other studies, whole expansins from various microbial sources were similarly found to bind to various forms of cellulose, xylan and lignocellulosic materials [8, 9, 10, 11, 12] but the contribution of the CBM63 domain was not ascertained (see review in [3]). Studies of native expansins from plant sources document binding to disordered cellulose by an α-expansin from cucumber hypocotyls (CsEXPA1, GenBank: KGN50732.1) [13] or to xylans by a β-expansin (ZmEXPB1, GenBank: NP_001288510.1) from maize pollen [14, 15]. Binding of ZmEXPB1 to cellulose (Avicel) in isolation is weaker than binding of BsEXLX1 (KD 5.8 μM, Bmax 0.4 μmol/g; ratio: 0.07 L/g versus 0.14 L/g for BsEXLX1), whereas it shows stronger binding to whole maize cell walls with complex kinetics that depend on buffer concentration [15]. NMR analysis of ZmEXPB1 targeting in whole maize cell walls shows that it interacts with matrix polysaccharides rather than cellulose [15]. This result is likely due to its greater affinity for the matrix components and to limited accessibility of cellulose in the complex cell wall. The specific contributions of the CBM63 domain to these binding interactions were not determined.
Structural Features
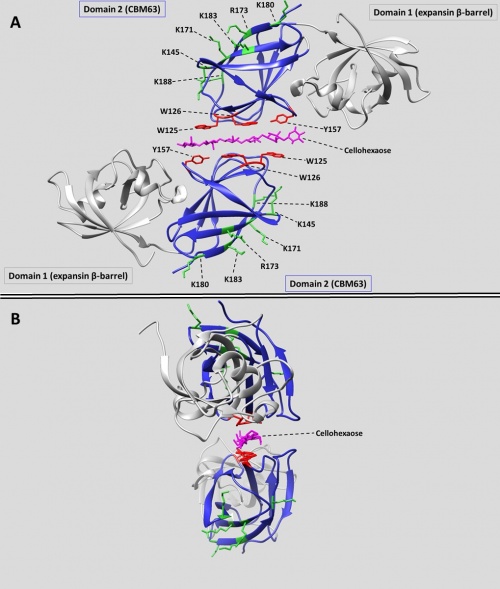
Crystal structures of CBM63s include two bacterial expansins (BsEXLX1 [5]: 3D30 and CmEXLX1 from Clavibacter michiganensis: 4JJO) and two plant expansins (ZmEXPB1 [14] from Zea mays: 2HCZ; and PhlP1 from Phleum pratense: 1N10). In addition, the Phleum pollen allergens PhlP2 (1WHO) and PhlP3 (3FT9) are evolutionarily derived from plant β-expansins and are homologous to CBM63 [16, 17], but their binding properties are unknown. The most convincing evidence that the C-terminal domain of plant expansins can be classified as CBM63 comes from structural alignments between the C-terminal domain of BsEXLX1 (the founding member of CBM63) and the C-terminal domain of the plant expansin ZmEXPB1. A structural superposition of these two domains showed a remarkably similar structure, with an RMSD of only 1.6 Å [5].
In BsEXLX1 the CBM63 fold consists of two sets of four anti-parallel β-strands that form a β-sandwich [5]. This fold forms a planar binding surface populated by three co-linear aromatic amino acids (W125, W126, Y157) which mediate hydrophobic binding interactions with cellohexaose and related cellulose-like oligosaccharides [6]. Thus, CBM63 is considered a type A CBM [11,12]. Mutagenesis of these residues reduces cellulose binding [7]. In crystal complexes with cello-oligosaccharides, BsEXLX1 forms an unusual structure in which a single glucan chain is sandwiched between two CBM63s from two different protein chains in opposite polarity (Figure 1-A). The aromatic residues of the two CBM63s interact with alternating glucose residues, always with the more hydrophobic face that is populated by three –CH groups. Hydrogen bonds are formed between the K119 residues of both proteins and several hydroxyl groups of cellohexaose. The cellohexaose bound in the sandwich structure displays a twist (Figure 1-B). Binding to cellulose is entropically driven due to the exclusion of bound water during the formation of CH-π interactions [6].
In the plant β-expansin ZmEXPB1, there are only two aromatic residues on the CBM63 surface (Y158, W192) [14], which may account for its weaker binding to cellulose. There are no crystal structures for alpha expansins, but sequence similarity and computational models suggest they have a similar 8-stranded β-sandwich fold [18].
Functionalities
BsEXLX1 induces creep of plant cell walls and reduces the breaking strength of filter paper strips but the CBM63 domain by itself lacks both of these activities [7]. In a BsEXLX1 mutant with reduced matrix binding, wall loosening increased, whereas a mutant with no binding to cellulose lost wall loosening activity, demonstrating that cell wall loosening activity of BsEXLX1 depends on binding to cellulose rather than the matrix [7]. Hence, matrix binding appears to be nonproductive for wall loosening by this protein. This conclusion is also supported by solid-state NMR studies [19] which showed that in partially-extracted cell walls from Arabidopsis, BsEXLX1 binds to a minor cellulose component with a chemical shift slightly different from the bulk of the cellulose. The implication of these results is that CBM63 targets expansins to specific sites, dubbed biomechanical hotspots [14, 15], where cell wall loosening occurs. It has been proposed that wall loosening requires the cooperative action of both expansin domains [7], but this idea needs additional testing. BsEXLX1 and other bacterial expansins have been tested for their ability to enhance cellulase action, but with mixed results (reviewed in [10]). Certain microbial expansins are fused to other CAZy domains, including CBM1, CBM2, and GH5 (summarized in [4]). These chimeric proteins presumably facilitate plant-microbe association (whether pathogenic or commensal), but the exact function and contribution of the CBM63 domain is unknown. Dockerin-fused expansins have been documented in Clostridium clariflavum (GenBank: WP_014255055.1), where they integrate into the cellulosome complex alongside other CBMs and endoglucanases [20]. Cellulosomal expansins bound microcrystalline cellulose [20], and the inclusion of expansin in native and designer cellulosomes enhanced cellulose degradation [20, 21], but again the exact contribution of CBM63 is unknown.
Family Firsts
- First Identified
- The isolation [11], characterization [13] and sequencing [12] of α-expansin led to speculation that the C-terminal region of expansin was a CBM-like domain [22].
- First Structural Characterization
- The crystal structures of ZmEXPB1 [14] and BsEXLX1 [5] confirmed the identity of the C-terminal domain as a type-A CBM, which was later recognized as a distinct family, dubbed CBM63 based on the C-terminal domain of BsEXLX1 from the soil bacterium Bacillus subtilis [6]. Crystal structural analysis of BsEXLX1 complexed with various cellulose-like oligosaccharides yielded the first crystal structure of a Type A CBM complexed with cellulose-like oligosaccharides [6].
References
- Cosgrove DJ (2015). Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol. 2015;25:162-72. DOI:10.1016/j.pbi.2015.05.014 |
- Cosgrove DJ (2016). Catalysts of plant cell wall loosening. F1000Res. 2016;5. DOI:10.12688/f1000research.7180.1 |
- Cosgrove DJ (2017). Microbial Expansins. Annu Rev Microbiol. 2017;71:479-497. DOI:10.1146/annurev-micro-090816-093315 |
- Nikolaidis N, Doran N, and Cosgrove DJ. (2014). Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol Biol Evol. 2014;31(2):376-86. DOI:10.1093/molbev/mst206 |
- Kerff F, Amoroso A, Herman R, Sauvage E, Petrella S, Filée P, Charlier P, Joris B, Tabuchi A, Nikolaidis N, and Cosgrove DJ. (2008). Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization. Proc Natl Acad Sci U S A. 2008;105(44):16876-81. DOI:10.1073/pnas.0809382105 |
- Georgelis N, Yennawar NH, and Cosgrove DJ. (2012). Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc Natl Acad Sci U S A. 2012;109(37):14830-5. DOI:10.1073/pnas.1213200109 |
- Georgelis N, Tabuchi A, Nikolaidis N, and Cosgrove DJ. (2011). Structure-function analysis of the bacterial expansin EXLX1. J Biol Chem. 2011;286(19):16814-23. DOI:10.1074/jbc.M111.225037 |
- Gilbert HJ, Knox JP, and Boraston AB. (2013). Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol. 2013;23(5):669-77. DOI:10.1016/j.sbi.2013.05.005 |
- Armenta S, Moreno-Mendieta S, Sánchez-Cuapio Z, Sánchez S, and Rodríguez-Sanoja R. (2017). Advances in molecular engineering of carbohydrate-binding modules. Proteins. 2017;85(9):1602-1617. DOI:10.1002/prot.25327 |
- Georgelis N, Nikolaidis N, and Cosgrove DJ. (2015). Bacterial expansins and related proteins from the world of microbes. Appl Microbiol Biotechnol. 2015;99(9):3807-23. DOI:10.1007/s00253-015-6534-0 |
- McQueen-Mason S, Durachko DM, and Cosgrove DJ. (1992). Two endogenous proteins that induce cell wall extension in plants. Plant Cell. 1992;4(11):1425-33. DOI:10.1105/tpc.4.11.1425 |
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, and Cosgrove DJ. (1995). Molecular cloning and sequence analysis of expansins--a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci U S A. 1995;92(20):9245-9. DOI:10.1073/pnas.92.20.9245 |
- McQueen-Mason SJ and Cosgrove DJ. (1995). Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 1995;107(1):87-100. DOI:10.1104/pp.107.1.87 |
- Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, and Cosgrove DJ. (2006). Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc Natl Acad Sci U S A. 2006;103(40):14664-71. DOI:10.1073/pnas.0605979103 |
- Wang T, Chen Y, Tabuchi A, Cosgrove DJ, and Hong M. (2016). The Target of β-Expansin EXPB1 in Maize Cell Walls from Binding and Solid-State NMR Studies. Plant Physiol. 2016;172(4):2107-2119. DOI:10.1104/pp.16.01311 |
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, and McQueen-Mason SJ. (2002). Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002;128(3):854-64. DOI:10.1104/pp.010658 |
- Sampedro J and Cosgrove DJ. (2005). The expansin superfamily. Genome Biol. 2005;6(12):242. DOI:10.1186/gb-2005-6-12-242 |
- Gaete-Eastman C, Morales-Quintana L, Herrera R, and Moya-León MA. (2015). In-silico analysis of the structure and binding site features of an α-expansin protein from mountain papaya fruit (VpEXPA2), through molecular modeling, docking, and dynamics simulation studies. J Mol Model. 2015;21(5):115. DOI:10.1007/s00894-015-2656-7 |
- Wang T, Park YB, Caporini MA, Rosay M, Zhong L, Cosgrove DJ, and Hong M. (2013). Sensitivity-enhanced solid-state NMR detection of expansin's target in plant cell walls. Proc Natl Acad Sci U S A. 2013;110(41):16444-9. DOI:10.1073/pnas.1316290110 |
- Artzi L, Morag E, Shamshoum M, and Bayer EA. (2016). Cellulosomal expansin: functionality and incorporation into the complex. Biotechnol Biofuels. 2016;9:61. DOI:10.1186/s13068-016-0474-5 |
- Chen C, Cui Z, Song X, Liu YJ, Cui Q, and Feng Y. (2016). Integration of bacterial expansin-like proteins into cellulosome promotes the cellulose degradation. Appl Microbiol Biotechnol. 2016;100(5):2203-12. DOI:10.1007/s00253-015-7071-6 |
- Cosgrove DJ (1996). Plant cell enlargement and the action of expansins. Bioessays. 1996;18(7):533-40. DOI:10.1002/bies.950180704 |