CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article.
Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 186"
Sei Motouchi (talk | contribs) |
Sei Motouchi (talk | contribs) |
||
| Line 30: | Line 30: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | The defining member of GH186, a β-1,2- | + | The defining member of GH186, a β-1,2-glucanse from ''Escherichia coli'' (EcOpgD) was identified, characterized and structurally analyzed as reported in 2023<cite>Motouchi2023</cite>. Subsequently GH186 homolog from ''Xanthomonas campestris'' pv. ''campestris'' (XccOpgD) was found to be anomer-inverting transglycoslyase which specifically substitute β-1,2-glucosidic bond of β-1,2-glucan to α-1,6-glucosidic bond at <cite>Motouchi2024</cite>. EcOpgD and XccOpgD are specific toward β-1,2-glucan and the amino acid residues for recognizing β-1,2-glucan at common subsites between EcOpgD and XccOpgD are highly conserved in GH186<cite>Motouchi2023 Motouchi2024</cite>. However, the reaction types of EcOpgD and XccOpgD are different from each other<cite>Motouchi2023 Motouchi2024</cite>. EcOpgD is a β-1,2-glucanase and preferentially generate β-1,2-glucooligosaccharides (Sopns, n is degree of polymerization, DP) with DPs of 6 and 7 from linear β-1,2-glucan<cite>Motouchi2023</cite>. Final products produced by EcOpgD are Sop6–10, indicating that EcOgpD hydrolyzes Sopns with DPs of 11 and higher<cite>Motouchi2023</cite>. XccOpgD generates only α-1,6-cyclized β-1,2-glucohexadecaose from linear β-1,2-glucan<cite>Motouchi2024</cite>. Almost all family members are found in Pseudomonadota, especially in gamma proteobacteria. Functionally important residues in EcOpgD and XccOpgD are not conserved in most of GH186 homologs<cite>Motouchi2023 Motouchi2024</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
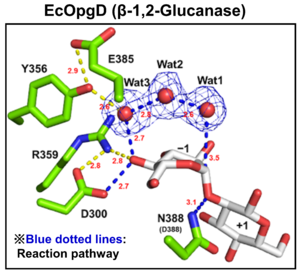
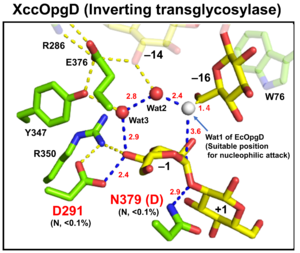
| − | [[File: | + | [[File:EcOgpD.png|thumb]][[File:XccOpgD.png|thumb]]Optical rotation and NMR analyses indicate that EcOpgD and XccOpgD adopt anomer-inverting mechanism<cite>Motouchi2023 Motouchi2024</cite>. X-ray structural analysis and mutational analysis suggest that D388 in EcOpgD and the equivalent residue in XccOpgD, D379, directly protonate the scissile glycosidic bond as general acids<cite>Motouchi2023 Motouchi2024</cite>. These analyses also suggest that D300 in EcOpgD and the equivalent residue in XccOpgD, D291, activate the nucleophile via 4-hydroxy group of the Glc moiety at subsite –1 and two water molecules as general bases<cite>Motouchi2023 Motouchi2024</cite>. Interestingly, nucleophiles of EcOpgD and XccOpgD are different from each other<cite>Motouchi2023 Motouchi2024</cite>. The nucleophiles of EcOpgD and XccOpgD are a water molecule and the 6-hydroxy group of Glc moiety at subsite –16, respectively<cite>Motouchi2023 Motouchi2024</cite>. The difference in nucleophile among GH186 family may be due to how a nucleophile and a following Grotthuss proton relay route are stabilized by residues and/or ligands. |
| + | |||
| + | Particularly, While W441 important for stabilizing nucleophilic water in EcOpgD is not conserved in GH186, W76 important for stacking acceptor Glc moiety in XccOpgD is broadly conserved in GH186 (but not in the clade of EcOpgD) in terms of the ability for stacking interaction. Therefore, GH186 seems fundamentally an anomer-inverting transglycosylase family. In addition, The Grotthuss proton relay pathway is sequestered (stabilized) not by amino acid sequence but by acceptor substrate moiety in XccOpgD, meaning that a water molecule is not suitable as nucleophile for efficient Grotthuss proton relay. This is probably the reason why XccOpgD is specific to transglycosylation. | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | General acid and base of EcOpgD are D388 and D300, respectively<cite> | + | General acid and base of EcOpgD are D388 and D300, respectively, and the catalytic residues of XccOpgD are also equivalent to that of EcOpgD (D379 and D291, respectively)<cite>Motouchi2023 Motouchi2024</cite>. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The ligand-free structure of OpgG from ''E. coli'' (EcOpgG) was determined at 2. | + | The ligand-free structure of OpgG from ''E. coli'' (EcOpgG) was determined at 2.4 Å (PDB: 1txk)<cite>Hanoulle2004</cite>. The ligand-free structure of EcOpgD was determined at 2.95 Å (PDB: 8IOX)<cite>Motouchi2023</cite>. Michaelis complexes of EcOpgD (D388N, co-crystal), EcOpgG (D361N, soaking) and XccOpgD (D379N, soaking) with β-1,2-glucan were determined at 2.06, 1.81, 2.25 Å, respectively (PDB: 8IP1, 8IP2, 8X18)<cite>Motouchi2023 Motouchi2024</cite>. |
| + | [[File:The overall structure of Michaelis complex of EcOpgG (monomer).jpg|thumb]]There is no structural homolog of GH186 in whole GH families<cite>Motouchi2023</cite> (January 2024). | ||
| + | |||
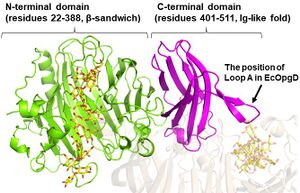
| + | EcOpgG consists of an N-terminal domain (residues 22–388, β-sandwich) and a C-terminal domain (residues 401–511, Ig-like fold). The two domains are connected with one turn of 3<sub>10</sub> helix<cite>Hanoulle2004 Motouchi2023</cite>. The loop region (residues 409-425, Loop A below) in the C-terminal domain of the ligand-free structure changes into β-strands in the Michaelis complex structure. In the Michaelis complex, the β-strands reach for the catalytic center of another chain in the dimer to cover the proton transfer pathway from a nucleophile to the general base catalyst<cite>Motouchi2023</cite>. However, the sequence of Loop A is diversified in GH186 family. Indeed, Loop A in EcOpgD sequesters the proton transfer pathway from the solvent, while that of EcOpgG does not completely, which is consistent with the drastically reduced hydrolytic activity of EcOpgG compared with EcOpgD<cite>Motouchi2023</cite>.In addition, the Loop A of XccOpgD is too short to reach the catalytic center, making the space for recoginizing acceptor β-1,2-glucooligosaccharide moiety<cite>Motouchi2024</cite>. | ||
== Family Firsts == | == Family Firsts == | ||
;First stereochemistry determination: EcOpgD by optical rotation<cite>MotouchiEc2023</cite>. | ;First stereochemistry determination: EcOpgD by optical rotation<cite>MotouchiEc2023</cite>. | ||
| Line 42: | Line 47: | ||
;First general base residue identification: EcOpgD by X-ray crystallography and site-directed mutagenesis<cite>MotouchiEc2023</cite>. | ;First general base residue identification: EcOpgD by X-ray crystallography and site-directed mutagenesis<cite>MotouchiEc2023</cite>. | ||
;First 3-D structure: EcOpgG by X-ray crystallography<cite>Hanoulle2004</cite>. | ;First 3-D structure: EcOpgG by X-ray crystallography<cite>Hanoulle2004</cite>. | ||
| − | |||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Motouchi2023 pmid=37735577 |
| + | #Motouchi2024 pmid=38957137 | ||
#Hanoulle2004 pmid=15313617 | #Hanoulle2004 pmid=15313617 | ||
</biblio> | </biblio> | ||
| − | |||
<!-- Do not delete this Category tag --> | <!-- Do not delete this Category tag --> | ||
[[Category:Glycoside Hydrolase Families|GH186]] | [[Category:Glycoside Hydrolase Families|GH186]] | ||
Revision as of 23:54, 4 July 2024
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| Glycoside Hydrolase Family GH186 | |
| Clan | GH-x |
| Mechanism | inverting |
| Active site residues | Asp |
| CAZy DB link | |
| http://www.cazy.org/GH186.html | |
Substrate specificities
The defining member of GH186, a β-1,2-glucanse from Escherichia coli (EcOpgD) was identified, characterized and structurally analyzed as reported in 2023[1]. Subsequently GH186 homolog from Xanthomonas campestris pv. campestris (XccOpgD) was found to be anomer-inverting transglycoslyase which specifically substitute β-1,2-glucosidic bond of β-1,2-glucan to α-1,6-glucosidic bond at [2]. EcOpgD and XccOpgD are specific toward β-1,2-glucan and the amino acid residues for recognizing β-1,2-glucan at common subsites between EcOpgD and XccOpgD are highly conserved in GH186[1, 2]. However, the reaction types of EcOpgD and XccOpgD are different from each other[1, 2]. EcOpgD is a β-1,2-glucanase and preferentially generate β-1,2-glucooligosaccharides (Sopns, n is degree of polymerization, DP) with DPs of 6 and 7 from linear β-1,2-glucan[1]. Final products produced by EcOpgD are Sop6–10, indicating that EcOgpD hydrolyzes Sopns with DPs of 11 and higher[1]. XccOpgD generates only α-1,6-cyclized β-1,2-glucohexadecaose from linear β-1,2-glucan[2]. Almost all family members are found in Pseudomonadota, especially in gamma proteobacteria. Functionally important residues in EcOpgD and XccOpgD are not conserved in most of GH186 homologs[1, 2].
Kinetics and Mechanism
Optical rotation and NMR analyses indicate that EcOpgD and XccOpgD adopt anomer-inverting mechanism[1, 2]. X-ray structural analysis and mutational analysis suggest that D388 in EcOpgD and the equivalent residue in XccOpgD, D379, directly protonate the scissile glycosidic bond as general acids[1, 2]. These analyses also suggest that D300 in EcOpgD and the equivalent residue in XccOpgD, D291, activate the nucleophile via 4-hydroxy group of the Glc moiety at subsite –1 and two water molecules as general bases[1, 2]. Interestingly, nucleophiles of EcOpgD and XccOpgD are different from each other[1, 2]. The nucleophiles of EcOpgD and XccOpgD are a water molecule and the 6-hydroxy group of Glc moiety at subsite –16, respectively[1, 2]. The difference in nucleophile among GH186 family may be due to how a nucleophile and a following Grotthuss proton relay route are stabilized by residues and/or ligands.
Particularly, While W441 important for stabilizing nucleophilic water in EcOpgD is not conserved in GH186, W76 important for stacking acceptor Glc moiety in XccOpgD is broadly conserved in GH186 (but not in the clade of EcOpgD) in terms of the ability for stacking interaction. Therefore, GH186 seems fundamentally an anomer-inverting transglycosylase family. In addition, The Grotthuss proton relay pathway is sequestered (stabilized) not by amino acid sequence but by acceptor substrate moiety in XccOpgD, meaning that a water molecule is not suitable as nucleophile for efficient Grotthuss proton relay. This is probably the reason why XccOpgD is specific to transglycosylation.
Catalytic Residues
General acid and base of EcOpgD are D388 and D300, respectively, and the catalytic residues of XccOpgD are also equivalent to that of EcOpgD (D379 and D291, respectively)[1, 2].
Three-dimensional structures
The ligand-free structure of OpgG from E. coli (EcOpgG) was determined at 2.4 Å (PDB: 1txk)[3]. The ligand-free structure of EcOpgD was determined at 2.95 Å (PDB: 8IOX)[1]. Michaelis complexes of EcOpgD (D388N, co-crystal), EcOpgG (D361N, soaking) and XccOpgD (D379N, soaking) with β-1,2-glucan were determined at 2.06, 1.81, 2.25 Å, respectively (PDB: 8IP1, 8IP2, 8X18)[1, 2].
There is no structural homolog of GH186 in whole GH families[1] (January 2024).
EcOpgG consists of an N-terminal domain (residues 22–388, β-sandwich) and a C-terminal domain (residues 401–511, Ig-like fold). The two domains are connected with one turn of 310 helix[1, 3]. The loop region (residues 409-425, Loop A below) in the C-terminal domain of the ligand-free structure changes into β-strands in the Michaelis complex structure. In the Michaelis complex, the β-strands reach for the catalytic center of another chain in the dimer to cover the proton transfer pathway from a nucleophile to the general base catalyst[1]. However, the sequence of Loop A is diversified in GH186 family. Indeed, Loop A in EcOpgD sequesters the proton transfer pathway from the solvent, while that of EcOpgG does not completely, which is consistent with the drastically reduced hydrolytic activity of EcOpgG compared with EcOpgD[1].In addition, the Loop A of XccOpgD is too short to reach the catalytic center, making the space for recoginizing acceptor β-1,2-glucooligosaccharide moiety[2].
Family Firsts
- First stereochemistry determination
- EcOpgD by optical rotation[4].
- First general acid residue identification
- EcOpgD by X-ray crystallography and site-directed mutagenesis[4].
- First general base residue identification
- EcOpgD by X-ray crystallography and site-directed mutagenesis[4].
- First 3-D structure
- EcOpgG by X-ray crystallography[3].
References
- Motouchi S, Kobayashi K, Nakai H, and Nakajima M. (2023). Identification of enzymatic functions of osmo-regulated periplasmic glucan biosynthesis proteins from Escherichia coli reveals a novel glycoside hydrolase family. Commun Biol. 2023;6(1):961. DOI:10.1038/s42003-023-05336-6 |
- Motouchi S, Komba S, Nakai H, and Nakajima M. (2024). Discovery of Anomer-Inverting Transglycosylase: Cyclic Glucohexadecaose-Producing Enzyme from Xanthomonas, a Phytopathogen. J Am Chem Soc. 2024;146(26):17738-17746. DOI:10.1021/jacs.4c02579 |
- Hanoulle X, Rollet E, Clantin B, Landrieu I, Odberg-Ferragut C, Lippens G, Bohin JP, and Villeret V. (2004). Structural analysis of Escherichia coli OpgG, a protein required for the biosynthesis of osmoregulated periplasmic glucans. J Mol Biol. 2004;342(1):195-205. DOI:10.1016/j.jmb.2004.07.004 |