CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 93"
Harry Brumer (talk | contribs) |
|||
| (83 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | * [[Author]]: Annabelle Varrot | + | {{CuratorApproved}} |
| − | * [[Responsible Curator]]: Annabelle Varrot | + | * [[Author]]: [[User:Annabelle Varrot|Annabelle Varrot]] |
| + | * [[Responsible Curator]]: [[User:Annabelle Varrot|Annabelle Varrot]] | ||
<div style="float:right"> | <div style="float:right"> | ||
| Line 18: | Line 19: | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
|- | |- | ||
| − | | colspan="2" | | + | | colspan="2" |{{CAZyDBlink}}GH93.html |
|} | |} | ||
</div> | </div> | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | The characterized | + | The characterized [[glycoside hydrolases]] of family GH93 are known to hydrolyse linear α-1,5-L-arabinan. <cite>Sakamoto2001 Carapito2009</cite>, EC:3.2.1-. |
| − | EC:3.2.1- | ||
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
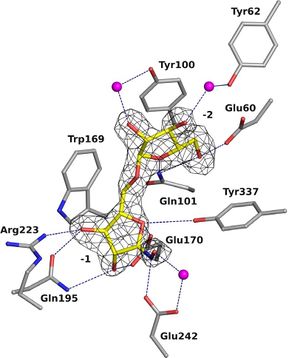
| − | GH93 enzymes are | + | [[Image:overallsurf.png|thumb|300px|right|'''Figure 1. Cartoon representation of the overall structure of Arb93A''' (PDB ID [{{PDBlink}}2w5o 2W5O] <cite>Carapito2009</cite>.The propeller is colored blue to red from the N- to C-terminus and arabinobiose is depicted as balls and sticks.]] |
| + | |||
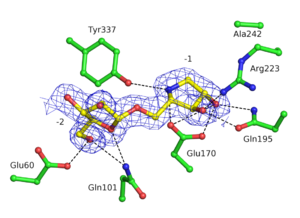
| + | GH93 enzymes are [[exo]]-acting enzymes that only release arabinobiose from the non-reducing end of α-1,5-L-arabinan. These enzymes are proposed to be [[retaining]] enzymes based on the net retention of the configuration of the anomeric carbon is proposed from the products of the transglycosylation activity of the protein Abnx from ''Penicillium chrysogenum'' <cite>Sakamoto2004</cite>. This proposal obtained support from the crystal structures of the Arb93A enzyme from ''Fusarium graminearum'' and Abnx both in complex with arabinobiose (Fig. 1) <cite>Carapito2009 Sogabe2011</cite>. α-L-Arabinofuranosylated pyrrolidines were shown to be good inhibitors of Arb93A. The Arb93A complex structure with a deoxyiminosugar equivalent of arabinobiose revealed a <sup>4</sup>T<sub>N</sub> twist conformation expected for the Michaelis complex, as seen for several retaining GH51 α-L-arabinofuranosidases (Fig. 2) <cite>GoddardBorger2011</cite>. Potent shape mimic inhibitors exploiting sp<sup>2</sup> hybridization at the anomeric carbon have been recently synthetized as well as a chromogenic substrate (Fig. 3). They are useful tools to assist further biochemical studies on L-arabinanases <cite>Coyle2017</cite>. | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | From the crystal structure of Arb93A, Glu170 and Glu242 are proposed to act as nucleophile and acid/base respectively. Mutagenesis experiment support their role in catalysis and they are strictly conserved | + | From the crystal structure of Arb93A, Glu170 and Glu242 are proposed to act as [[catalytic nucleophile]] and [[general acid/base]] respectively. Mutagenesis experiment support their role in catalysis and they are strictly conserved among the family members <cite>Carapito2009</cite>. Recent structures and mutagenesis studies for the arabinanase Abnx from ''Penicillium chrysogenum 31B'' strengthened this assignment. Mutations to alanine or glutamine of their equivalent Glu174 and Glu246 lead to inactive enzyme <cite>Sogabe2011</cite>. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The | + | [[Image:242den.png|thumb|right|300px|'''Figure 2. Electron density for deoxyiminoarabinobiose bound to the active site of Arb93A (PDB ID [{{PDBlink}}2ydt 2YDT] <cite>GoddardBorger2011</cite>.''' Hydrogens bond are represented as dashed lines.]] |
| − | of sialidases of clan | + | [[File:62 araf chembiochem-17.jpg|thumb|right|300px|'''Figure 3. Electron density for hydroximolactone inhibitor bound to the active site of Arb93A (PDB ID [{{PDBlink}}5m1z 5M1Z].<cite>Coyle2017</cite>''']] |
| − | + | ||
| + | The crystal structure of Arb93A reveals a six-bladed β-propeller fold characteristic of [[GH33]], [[GH34]], and [[GH83]] sialidases, which are also members of [[clan]] GH-E (Fig. 3) <cite>Carapito2009 Sogabe2011</cite>. The catalytic machinery is however very different from that of sialidases <cite>Gaskell1995</cite>. The wild-type structure was solved at 2.05 angstrom resolution in complex with arabinobiose . The active site is located in a deep acidic L-shaped crevice at the center of the beta-propeller (Fig. 1). Structures of the wild-type or E242A mutant enzyme in complex with iminoarabinobiose were solved at 1.6 and 1.85 angstrom resolution respectively as well as a complex with a shape mimic inhibitor and demonstrated ring distorsion (Fig. 2-3) <cite>GoddardBorger2011 Coyle2017</cite>. | ||
== Family Firsts == | == Family Firsts == | ||
| + | ;First sterochemistry determination: This was determined with the ''Penicillium chrysogenum'' Abxn enzyme using <sup>1</sup>H-NMR to identify the transglycosylation products <cite>Sakamoto2004</cite> | ||
| + | ;First [[catalytic nucleophile]] identification: This was proposed based on the structure of ''Fusarium graminearum'' Arb93A <cite>Carapito2009</cite> | ||
| + | ;First [[general acid/base]] residue identification: This was proposed based on the structure of ''Fusarium graminearum'' Arb93A <cite>Carapito2009</cite> | ||
| + | ;First 3-D structure: Determined for ''Fusarium graminearum'' Arb93A by Carapito and co-workers <cite>Carapito2009</cite> | ||
| + | == References == | ||
| + | <biblio> | ||
| − | == | + | #Sakamoto2001 pmid=11425761 |
| + | #Carapito2009 pmid=19269961 | ||
| + | #Sakamoto2004 pmid=15342117 | ||
| + | #Sogabe2011 pmid=21543843 | ||
| + | #GoddardBorger2011 pmid=21773614 | ||
| + | #Coyle2017 pmid=28266777 | ||
| + | #Gaskell1995 pmid=8591030 | ||
| + | </biblio> | ||
| + | |||
| + | <!-- DO NOT REMOVE THIS CATEGORY TAG! (...but please delete the nowiki tags before saving.) --> | ||
| + | [[Category:Glycoside Hydrolase Families|GH093]] | ||
Latest revision as of 08:17, 5 March 2019
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH93 | |
| Clan | GH-E |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH93.html | |
Substrate specificities
The characterized glycoside hydrolases of family GH93 are known to hydrolyse linear α-1,5-L-arabinan. [1, 2], EC:3.2.1-.
Kinetics and Mechanism
GH93 enzymes are exo-acting enzymes that only release arabinobiose from the non-reducing end of α-1,5-L-arabinan. These enzymes are proposed to be retaining enzymes based on the net retention of the configuration of the anomeric carbon is proposed from the products of the transglycosylation activity of the protein Abnx from Penicillium chrysogenum [3]. This proposal obtained support from the crystal structures of the Arb93A enzyme from Fusarium graminearum and Abnx both in complex with arabinobiose (Fig. 1) [2, 4]. α-L-Arabinofuranosylated pyrrolidines were shown to be good inhibitors of Arb93A. The Arb93A complex structure with a deoxyiminosugar equivalent of arabinobiose revealed a 4TN twist conformation expected for the Michaelis complex, as seen for several retaining GH51 α-L-arabinofuranosidases (Fig. 2) [5]. Potent shape mimic inhibitors exploiting sp2 hybridization at the anomeric carbon have been recently synthetized as well as a chromogenic substrate (Fig. 3). They are useful tools to assist further biochemical studies on L-arabinanases [6].
Catalytic Residues
From the crystal structure of Arb93A, Glu170 and Glu242 are proposed to act as catalytic nucleophile and general acid/base respectively. Mutagenesis experiment support their role in catalysis and they are strictly conserved among the family members [2]. Recent structures and mutagenesis studies for the arabinanase Abnx from Penicillium chrysogenum 31B strengthened this assignment. Mutations to alanine or glutamine of their equivalent Glu174 and Glu246 lead to inactive enzyme [4].
Three-dimensional structures
The crystal structure of Arb93A reveals a six-bladed β-propeller fold characteristic of GH33, GH34, and GH83 sialidases, which are also members of clan GH-E (Fig. 3) [2, 4]. The catalytic machinery is however very different from that of sialidases [7]. The wild-type structure was solved at 2.05 angstrom resolution in complex with arabinobiose . The active site is located in a deep acidic L-shaped crevice at the center of the beta-propeller (Fig. 1). Structures of the wild-type or E242A mutant enzyme in complex with iminoarabinobiose were solved at 1.6 and 1.85 angstrom resolution respectively as well as a complex with a shape mimic inhibitor and demonstrated ring distorsion (Fig. 2-3) [5, 6].
Family Firsts
- First sterochemistry determination
- This was determined with the Penicillium chrysogenum Abxn enzyme using 1H-NMR to identify the transglycosylation products [3]
- First catalytic nucleophile identification
- This was proposed based on the structure of Fusarium graminearum Arb93A [2]
- First general acid/base residue identification
- This was proposed based on the structure of Fusarium graminearum Arb93A [2]
- First 3-D structure
- Determined for Fusarium graminearum Arb93A by Carapito and co-workers [2]
References
- Sakamoto T and Thibault JF. (2001). Exo-arabinanase of Penicillium chrysogenum able to release arabinobiose from alpha-1,5-L-arabinan. Appl Environ Microbiol. 2001;67(7):3319-21. DOI:10.1128/AEM.67.7.3319-3321.2001 |
- Carapito R, Imberty A, Jeltsch JM, Byrns SC, Tam PH, Lowary TL, Varrot A, and Phalip V. (2009). Molecular basis of arabinobio-hydrolase activity in phytopathogenic fungi: crystal structure and catalytic mechanism of Fusarium graminearum GH93 exo-alpha-L-arabinanase. J Biol Chem. 2009;284(18):12285-96. DOI:10.1074/jbc.M900439200 |
- Sakamoto T, Fujita T, and Kawasaki H. (2004). Transglycosylation catalyzed by a Penicillium chrysogenum exo-1,5-alpha-L-arabinanase. Biochim Biophys Acta. 2004;1674(1):85-90. DOI:10.1016/j.bbagen.2004.06.001 |
- Sogabe Y, Kitatani T, Yamaguchi A, Kinoshita T, Adachi H, Takano K, Inoue T, Mori Y, Matsumura H, Sakamoto T, and Tada T. (2011). High-resolution structure of exo-arabinanase from Penicillium chrysogenum. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 5):415-22. DOI:10.1107/S0907444911006299 |
- Goddard-Borger ED, Carapito R, Jeltsch JM, Phalip V, Stick RV, and Varrot A. (2011). α-L-Arabinofuranosylated pyrrolidines as arabinanase inhibitors. Chem Commun (Camb). 2011;47(34):9684-6. DOI:10.1039/c1cc13675e |
- Coyle T, Debowski AW, Varrot A, and Stubbs KA. (2017). Exploiting sp(2) -Hybridisation in the Development of Potent 1,5-α-l-Arabinanase Inhibitors. Chembiochem. 2017;18(11):974-978. DOI:10.1002/cbic.201700073 |
- Gaskell A, Crennell S, and Taylor G. (1995). The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3(11):1197-205. DOI:10.1016/s0969-2126(01)00255-6 |