CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 136"
| Line 43: | Line 43: | ||
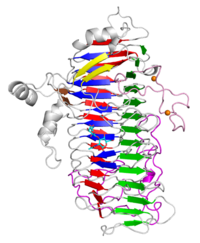
The X-ray crystal structure of the catalytic domain, LnbXc(31-625) revealed a right-handed β helix fold. | The X-ray crystal structure of the catalytic domain, LnbXc(31-625) revealed a right-handed β helix fold. | ||
Three forms, ligand free (2.36 Å resolution), LNB complex (1.82 Å), and GNB complex (2.70 Å) were determined. | Three forms, ligand free (2.36 Å resolution), LNB complex (1.82 Å), and GNB complex (2.70 Å) were determined. | ||
| − | [[ | + | [[file:LnbXc.png|200px|right]] |
== Family Firsts == | == Family Firsts == | ||
Revision as of 21:04, 12 August 2020
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Chihaya Yamada^^^
- Responsible Curator: ^^^Shinya Fushinobu^^^
| Glycoside Hydrolase Family GH136 | |
| Clan | GH-N |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH136.html | |
Substrate specificities
This family of glycoside hydrolases contains lacto-N-biosidase, as demonstrated for LnbX from Bifidobacterium longum JCM 1217 [1]. LnbX liberates Galβ1-3GlcNAc(lacto-N-biose I, LNB) and lactose from lacto-N-tetraose, the main component of human milk oligosaccharides. It hydrolyzed the linkage GlcNAcβ1-3Gal in lacto-N-hexaose, lacto-N-fucopentaose I, and sialyllacto-N-tetraose a of human milk oligosaccharides as substrate of LnbX in the GH136. In addition, LnbX liberates Galβ1-3GalNAc (GNB) from the sugar chains of globo- and ganglio-series glycosphingolipids [2].
GH136 lacto-N-biosidase required neighboring chaperon gene for folding. Rarely, chaperone-like gene fused to lacto-N-biosidase gene in case of ErGH136I and ErGH136IIfrom Eubacterium ramulus [3].
Kinetics and Mechanism
Content is to be added here.
Catalytic Residues
The nucleophile is Asp418. The catalytic acid/base is Asp411 via water molecule.
Three-dimensional structures
The X-ray crystal structure of the catalytic domain, LnbXc(31-625) revealed a right-handed β helix fold. Three forms, ligand free (2.36 Å resolution), LNB complex (1.82 Å), and GNB complex (2.70 Å) were determined.
Family Firsts
- First stereochemistry determination
- Content is to be added here.
- First catalytic nucleophile identification
- Content is to be added here.
- First general acid/base residue identification
- Content is to be added here.
- First 3-D structure
- Content is to be added here.
References
- Sakurama H, Kiyohara M, Wada J, Honda Y, Yamaguchi M, Fukiya S, Yokota A, Ashida H, Kumagai H, Kitaoka M, Yamamoto K, and Katayama T. (2013). Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J Biol Chem. 2013;288(35):25194-25206. DOI:10.1074/jbc.M113.484733 |
- Gotoh A, Katoh T, Sugiyama Y, Kurihara S, Honda Y, Sakurama H, Kambe T, Ashida H, Kitaoka M, Yamamoto K, and Katayama T. (2015). Novel substrate specificities of two lacto-N-biosidases towards β-linked galacto-N-biose-containing oligosaccharides of globo H, Gb5, and GA1. Carbohydr Res. 2015;408:18-24. DOI:10.1016/j.carres.2015.03.005 |
- Pichler MJ, Yamada C, Shuoker B, Alvarez-Silva C, Gotoh A, Leth ML, Schoof E, Katoh T, Sakanaka M, Katayama T, Jin C, Karlsson NG, Arumugam M, Fushinobu S, and Abou Hachem M. (2020). Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat Commun. 2020;11(1):3285. DOI:10.1038/s41467-020-17075-x |
- Yamada C, Gotoh A, Sakanaka M, Hattie M, Stubbs KA, Katayama-Ikegami A, Hirose J, Kurihara S, Arakawa T, Kitaoka M, Okuda S, Katayama T, and Fushinobu S. (2017). Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum. Cell Chem Biol. 2017;24(4):515-524.e5. DOI:10.1016/j.chembiol.2017.03.012 |