CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Auxiliary Activity Family 13"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (18 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Glyn Hemsworth|Glyn Hemsworth]] & [[User:Leila LoLeggio|Leila LoLeggio]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Gideon Davies|Gideon Davies]] |
---- | ---- | ||
<!-- The data in the table below should be updated by the Author/Curator according to current information on the family --> | <!-- The data in the table below should be updated by the Author/Curator according to current information on the family --> | ||
<div style="float:right"> | <div style="float:right"> | ||
| − | {| {{Prettytable}} | + | {| {{Prettytable}} |
|- | |- | ||
|{{Hl2}} colspan="2" align="center" |'''Auxiliary Activity Family 13''' | |{{Hl2}} colspan="2" align="center" |'''Auxiliary Activity Family 13''' | ||
| Line 29: | Line 29: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | The AA13 family represents the fourth family of Lytic Polysaccharide Monooxygenases (LPMOs) that has been identified. Found in fungi, the first member of this family to be identified and characterised in the academic literature was isolated from <i>Neurospora crassa</i> by Vu et al <cite>Vu2014</cite>. | + | The AA13 family represents the fourth family of (Lytic) Polysaccharide Monooxygenases (LPMOs / sometimes called PMOs) that has been identified. Found in fungi, the first member of this family to be identified and characterised in the academic literature was isolated from <i>Neurospora crassa</i> by Vu et al <cite>Vu2014</cite>. While searching the <i>Neurospora crassa</i> genome for new putative LPMO sequences, they identified a sequence that appeared to code for an LPMO with a C-terminal [[CBM20]] domain but lacked significant overall sequence similarity to other [[AA9]], [[AA10]] or [[AA11]] LPMOs. Subsequently, Lo Leggio et al <cite>LoLeggio2015</cite> also identified and characterised AA13 family members from <i>Aspergilllus nidulans</i> and <i>Aspergillus oryzae</i> using a similar approach. Prior to this, genes now known to code for AA13s were demonstrated to produce proteins capable of boosting α and β-amylase activity during starch, amylose and amylopectin degradation ([{{PatentLink}}WO2014197705A1 WO2014197705A1]). |
| − | + | Of the AA13s that have been biochemically characterised to date, activity has only been demonstrate on starch and related substrates <cite>Vu2014 LoLeggio2015</cite>. The expression of these enzymes has also been shown to be heavily unregulated during growth of <i>A. nidulans</i> on starch <cite>Nekiunaite2016a</cite>. As for other LPMOs, AA13s utilise copper and an electron donor to oxidatively introduce chain breaks into the α-1,4-linked glucose polymers that form starch <cite>Vu2014 LoLeggio2015</cite>. AA13s specifically attack at the C1 position of the sugar ring forming oligosaccharide products with lactones at the reducing end which are then additionally hydrated to form aldonic acid terminated maltodextrins <cite>LoLeggio2015</cite>. The C1 specificity suggests that these LPMOs should be able to oxidatively attack both the α-1,4- and α-1,6-linkages found in amylopectin <cite>Vu2015</cite>. Interestingly, AA13s are currently the only LPMOs characterised to date that act on a glucan polymer formed from anything other than β-1,4-linkages <cite>Vu2016</cite>. | |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | All LPMOs ([[AA9]], [[AA10]], [[AA11]], & AA13) are copper dependent monooxygenases and there is considerable work ongoing in trying to elucidate their detailed reaction mechanism <cite>Walton2017 | + | All LPMOs ([[AA9]], [[AA10]], [[AA11]], & AA13) are copper dependent monooxygenases and there is considerable work ongoing in trying to elucidate their detailed reaction mechanism <cite>Walton2017</cite>. Starch, being an α-1,4-linked substrate, poses a different challenge for AA13s to be able to oxidatively attack at the C1 carbon. Whether there is a significant difference in mechanism required in order to oxidise starch is currently unclear. |
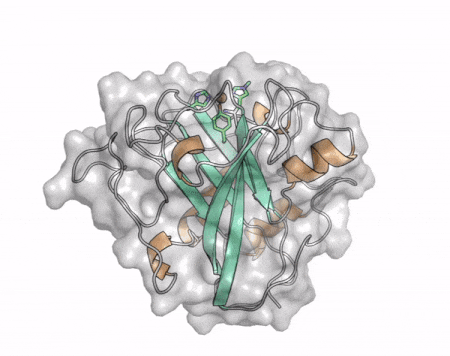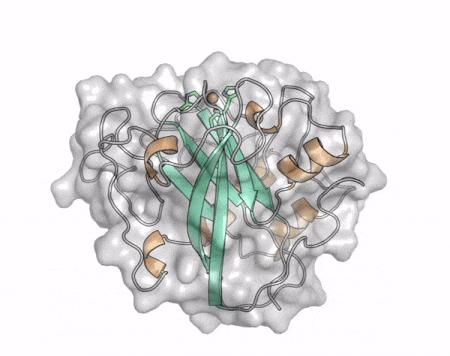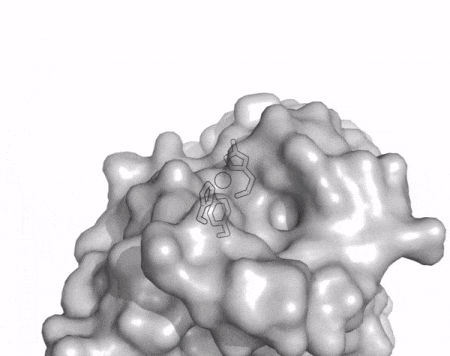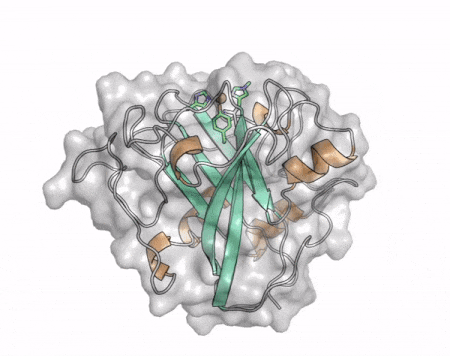
[[File:AA13_Cazypedia_Small.gif|thumb|right|450x356px|'''Figure 1. Structure of AA13 from <i>Aspergillus oryzae</i> (PDB ID [{{PDBlink}}4opb 4OPB] <cite>LoLeggio2015</cite>).''' In the above animation the structure is shown initially in cartoon representation coloured by secondary structure and the transparent surface shown in grey. The active site is shown at the top of the structure with the copper ion shown as a sphere and surrounding active site residues shown as sticks with green carbon atoms. As the animation proceeds the surface surrounding the active site is shown to illustrate the presence of a groove that passes over the active site. This groove is thought to accommodate the helical structure of an α-1,4-linked amylopectin substrate.]] | [[File:AA13_Cazypedia_Small.gif|thumb|right|450x356px|'''Figure 1. Structure of AA13 from <i>Aspergillus oryzae</i> (PDB ID [{{PDBlink}}4opb 4OPB] <cite>LoLeggio2015</cite>).''' In the above animation the structure is shown initially in cartoon representation coloured by secondary structure and the transparent surface shown in grey. The active site is shown at the top of the structure with the copper ion shown as a sphere and surrounding active site residues shown as sticks with green carbon atoms. As the animation proceeds the surface surrounding the active site is shown to illustrate the presence of a groove that passes over the active site. This groove is thought to accommodate the helical structure of an α-1,4-linked amylopectin substrate.]] | ||
| − | Performing kinetic measurements on LPMOs has been notoriously difficult. Several key aspects of AA13 biochemistry have been observed however. The <i>A. nidulans</i> AA13 showed significant synergy | + | Performing kinetic measurements on LPMOs has been notoriously difficult. Several key aspects of AA13 biochemistry have been observed however. The <i>A. nidulans</i> AA13 showed significant synergy with a β-amylase during degradation assays using retrograded starch as the substrate. The combined action of these enzymes resulted in >100-fold increase in the release of maltose compared to β-amylase acting alone, representing one of the most significant boosting activities observed so far for any LPMO <cite>LoLeggio2015</cite>. Also highlighted in this study was the importance of the reducing agent used to activate the enzyme with cysteine providing greater activity than ascorbate which is typically used to activate LPMOs. Vu et al <cite>Vu2014</cite> also showed that cellobiose dehydrogenase (CDH composed of [[AA3]] and [[AA8]] domains) represents a good activating partner for AA13s as has been observed for [[AA9]]s. The importance of the [[CBM20]] module for AA13 activity has also been highlighted. The <i>A. oryzae</i> AA13, which naturally lacks a CBM, did not appear to be active in assays on starch while the [[CBM20]] appended A.nidulans enzyme showed clear activity <cite>LoLeggio2015</cite>. Indeed the presence of the [[CBM20]] domain has been demonstrated to confer typical starch and β-cyclodextrin binding properties to AA13s and is suggested to mediate most of the substrate binding properties of this family of enzymes <cite>Nekiunaite2017b</cite>. |
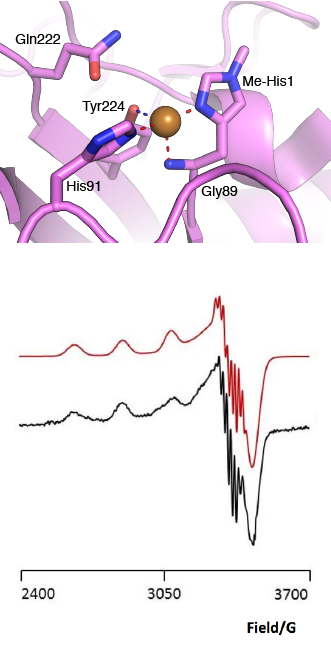
[[File:AA13_Active_Site.png|thumb|right|331x654px|'''Figure 2. The active site architecture observed for the AA13 from <i>A. oryzae</i> (PDB ID [{{PDBlink}}4opb 4OPB] <cite>LoLeggio2015</cite>).''' '''Top:''' the active site residues surrounding the copper ion are shown. The Cu is shown as a sphere and is in the Cu(I) state due to photo reduction from the X-ray beam. '''Bottom:''' EPR spectrum observed for the AA13 from <i>A. oryzae</i> revealing super hyperfine coupling. The raw data are shown in black with the simulated data in red.]] | [[File:AA13_Active_Site.png|thumb|right|331x654px|'''Figure 2. The active site architecture observed for the AA13 from <i>A. oryzae</i> (PDB ID [{{PDBlink}}4opb 4OPB] <cite>LoLeggio2015</cite>).''' '''Top:''' the active site residues surrounding the copper ion are shown. The Cu is shown as a sphere and is in the Cu(I) state due to photo reduction from the X-ray beam. '''Bottom:''' EPR spectrum observed for the AA13 from <i>A. oryzae</i> revealing super hyperfine coupling. The raw data are shown in black with the simulated data in red.]] | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The core fold of the AA13 structure is similar to all other LPMOs ([[AA9]], [[AA10]], & [[AA11]]) representing a β-sandwich immunoglobulin like fold <cite>LoLeggio2015</cite>. There are some significant structural differences compared to the other families however. Firstly, the AA13 structure reveals additional helical secondary structure elements that are not found in other LPMOs (Figure 1). The most prominent difference however is surrounding the copper active site. Where in other LPMOs, the copper histidine brace is found at the centre of a flat or slightly concave surface ([[AA9]], [[AA10]], & [[AA11]]), a distinct groove can be observed passing over the active site of AA13 (Figure 1). This is an adaptation that is thought to be necessary to accommodate the helical structure of an amylopectin substrate. | + | The core fold of the AA13 structure is similar to all other LPMOs ([[AA9]], [[AA10]], & [[AA11]]) representing a β-sandwich immunoglobulin like fold <cite>LoLeggio2015</cite>. There are some significant structural differences compared to the other families however. Firstly, the AA13 structure reveals additional helical secondary structure elements that are not found in other LPMOs (Figure 1). The most prominent difference however is surrounding the copper active site. Where in other LPMOs, the copper histidine brace is found at the centre of a flat or slightly concave surface ([[AA9]], [[AA10]], & [[AA11]]), a distinct groove can be observed passing over the active site of AA13 (Figure 1). This is an adaptation that is thought to be necessary to accommodate the helical structure of an amylopectin substrate. Despite significant efforts, it has not been possible to determine an oligosaccharide bound structure for an AA13 which would be particularly beneficial to delineating the adaptions necessary to allow activity on an α-1,4-linked substrate <cite>Frandsen2017</cite>. |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | The “histidine brace” motif is used to bind the active site copper in AA13 <cite>LoLeggio2015</cite>, as it is for all other LPMOs studied to date ([[AA9]], [[AA10]] & [[AA11]]) <cite>Vaaje-Kolstad2017</cite>. This motif sees the amino group and imidazole side chain of the N-terminal histidine, together with the imidazole group of a second histidine, directly coordinate the copper ion in a T-shaped geometry. For AA13s, one axial position is occupied by a tyrosine on one side of the copper ion, with a loop containing a glycine approaching near to the copper on the other side (Figure 2, top) <cite>LoLeggio2015</cite>. In [[AA10]]s and [[AA11]]s alanine in this position has been suggested as a possible important factor in distorting the arrangement of water molecules around the active site which may have mechanistic consequences in these families <cite> | + | The “histidine brace” motif is used to bind the active site copper in AA13 <cite>LoLeggio2015</cite>, as it is for all other LPMOs studied to date ([[AA9]], [[AA10]] & [[AA11]]) <cite>Vaaje-Kolstad2017</cite>. This motif sees the amino group and imidazole side chain of the N-terminal histidine, together with the imidazole group of a second histidine, directly coordinate the copper ion in a T-shaped geometry. For AA13s, one axial position is occupied by a tyrosine on one side of the copper ion, with a loop containing a glycine approaching near to the copper on the other side (Figure 2, top) <cite>LoLeggio2015</cite>. In [[AA10]]s and [[AA11]]s alanine in this position has been suggested as a possible important factor in distorting the arrangement of water molecules around the active site which may have mechanistic consequences in these families <cite>Hemsworth2013 Hemsworth2014</cite>. An interesting feature of the electron paramagnetic resonance (EPR) spectrum observed for the <i>A. nidulans</i> and <i>A. oryzae</i> AA13s was the presence of superhyperfine coupling in the spectrum (Figure 2, bottom) <cite>LoLeggio2015</cite>. |
== Family Firsts == | == Family Firsts == | ||
| Line 60: | Line 60: | ||
#Vu2016 pmid=27170366 | #Vu2016 pmid=27170366 | ||
#Walton2017 pmid=27094791 | #Walton2017 pmid=27094791 | ||
| − | |||
| − | |||
#Nekiunaite2017b pmid=27397613 | #Nekiunaite2017b pmid=27397613 | ||
#Vaaje-Kolstad2017 pmid=28086105 | #Vaaje-Kolstad2017 pmid=28086105 | ||
| − | # | + | #Frandsen2017 pmid=28045386 |
| − | |||
| − | |||
| − | |||
| + | #Hemsworth2013 pmid=23769965 | ||
| + | |||
| + | #Hemsworth2014 pmid=24362702 | ||
</biblio> | </biblio> | ||
| − | [[Category:Auxiliary Activity Families| | + | [[Category:Auxiliary Activity Families|AA013]] |
Latest revision as of 13:16, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Auxiliary Activity Family 13 | |
| Clan | Structurally related to AA9, AA10 & AA11 |
| Mechanism | lytic oxidase |
| Active site residues | mononuclear copper ion coordinated by the “histidine brace” |
| CAZy DB link | |
| https://www.cazy.org/AA13.html | |
Substrate specificities
The AA13 family represents the fourth family of (Lytic) Polysaccharide Monooxygenases (LPMOs / sometimes called PMOs) that has been identified. Found in fungi, the first member of this family to be identified and characterised in the academic literature was isolated from Neurospora crassa by Vu et al [1]. While searching the Neurospora crassa genome for new putative LPMO sequences, they identified a sequence that appeared to code for an LPMO with a C-terminal CBM20 domain but lacked significant overall sequence similarity to other AA9, AA10 or AA11 LPMOs. Subsequently, Lo Leggio et al [2] also identified and characterised AA13 family members from Aspergilllus nidulans and Aspergillus oryzae using a similar approach. Prior to this, genes now known to code for AA13s were demonstrated to produce proteins capable of boosting α and β-amylase activity during starch, amylose and amylopectin degradation (WO2014197705A1).
Of the AA13s that have been biochemically characterised to date, activity has only been demonstrate on starch and related substrates [1, 2]. The expression of these enzymes has also been shown to be heavily unregulated during growth of A. nidulans on starch [3]. As for other LPMOs, AA13s utilise copper and an electron donor to oxidatively introduce chain breaks into the α-1,4-linked glucose polymers that form starch [1, 2]. AA13s specifically attack at the C1 position of the sugar ring forming oligosaccharide products with lactones at the reducing end which are then additionally hydrated to form aldonic acid terminated maltodextrins [2]. The C1 specificity suggests that these LPMOs should be able to oxidatively attack both the α-1,4- and α-1,6-linkages found in amylopectin [4]. Interestingly, AA13s are currently the only LPMOs characterised to date that act on a glucan polymer formed from anything other than β-1,4-linkages [5].
Kinetics and Mechanism
All LPMOs (AA9, AA10, AA11, & AA13) are copper dependent monooxygenases and there is considerable work ongoing in trying to elucidate their detailed reaction mechanism [6]. Starch, being an α-1,4-linked substrate, poses a different challenge for AA13s to be able to oxidatively attack at the C1 carbon. Whether there is a significant difference in mechanism required in order to oxidise starch is currently unclear.
Performing kinetic measurements on LPMOs has been notoriously difficult. Several key aspects of AA13 biochemistry have been observed however. The A. nidulans AA13 showed significant synergy with a β-amylase during degradation assays using retrograded starch as the substrate. The combined action of these enzymes resulted in >100-fold increase in the release of maltose compared to β-amylase acting alone, representing one of the most significant boosting activities observed so far for any LPMO [2]. Also highlighted in this study was the importance of the reducing agent used to activate the enzyme with cysteine providing greater activity than ascorbate which is typically used to activate LPMOs. Vu et al [1] also showed that cellobiose dehydrogenase (CDH composed of AA3 and AA8 domains) represents a good activating partner for AA13s as has been observed for AA9s. The importance of the CBM20 module for AA13 activity has also been highlighted. The A. oryzae AA13, which naturally lacks a CBM, did not appear to be active in assays on starch while the CBM20 appended A.nidulans enzyme showed clear activity [2]. Indeed the presence of the CBM20 domain has been demonstrated to confer typical starch and β-cyclodextrin binding properties to AA13s and is suggested to mediate most of the substrate binding properties of this family of enzymes [7].
Three-dimensional structures
The core fold of the AA13 structure is similar to all other LPMOs (AA9, AA10, & AA11) representing a β-sandwich immunoglobulin like fold [2]. There are some significant structural differences compared to the other families however. Firstly, the AA13 structure reveals additional helical secondary structure elements that are not found in other LPMOs (Figure 1). The most prominent difference however is surrounding the copper active site. Where in other LPMOs, the copper histidine brace is found at the centre of a flat or slightly concave surface (AA9, AA10, & AA11), a distinct groove can be observed passing over the active site of AA13 (Figure 1). This is an adaptation that is thought to be necessary to accommodate the helical structure of an amylopectin substrate. Despite significant efforts, it has not been possible to determine an oligosaccharide bound structure for an AA13 which would be particularly beneficial to delineating the adaptions necessary to allow activity on an α-1,4-linked substrate [8].
Catalytic Residues
The “histidine brace” motif is used to bind the active site copper in AA13 [2], as it is for all other LPMOs studied to date (AA9, AA10 & AA11) [9]. This motif sees the amino group and imidazole side chain of the N-terminal histidine, together with the imidazole group of a second histidine, directly coordinate the copper ion in a T-shaped geometry. For AA13s, one axial position is occupied by a tyrosine on one side of the copper ion, with a loop containing a glycine approaching near to the copper on the other side (Figure 2, top) [2]. In AA10s and AA11s alanine in this position has been suggested as a possible important factor in distorting the arrangement of water molecules around the active site which may have mechanistic consequences in these families [10, 11]. An interesting feature of the electron paramagnetic resonance (EPR) spectrum observed for the A. nidulans and A. oryzae AA13s was the presence of superhyperfine coupling in the spectrum (Figure 2, bottom) [2].
Family Firsts
- First family member identified
- AA13 from N. crassa [1].
- First demonstration of oxidative cleavage
- The N. crassa AA13 was shown to produce oxidised malto-oligosaccharides in the presence of oxygen and reducing agents [1].
- First 3-D structure
- AA13 from A. oryzae with Cu+ 4OPB [2]
References
- Vu VV, Beeson WT, Span EA, Farquhar ER, and Marletta MA. (2014). A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci U S A. 2014;111(38):13822-7. DOI:10.1073/pnas.1408090111 |
- Lo Leggio L, Simmons TJ, Poulsen JC, Frandsen KE, Hemsworth GR, Stringer MA, von Freiesleben P, Tovborg M, Johansen KS, De Maria L, Harris PV, Soong CL, Dupree P, Tryfona T, Lenfant N, Henrissat B, Davies GJ, and Walton PH. (2015). Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun. 2015;6:5961. DOI:10.1038/ncomms6961 |
-
Nekiunaite, L., Arntzen, M.Ø., Svensson, B., Vaaje-Kolstad, G., Abou Hachem, M. (2016) Lytic polysaccharide monooxygenases and other oxidative enzymes are abundantly secreted by Aspergillus nidulans grown on different starches. Biotechnology for Biofuels. 9, 187 [1]
- Vu VV and Marletta MA. (2016). Starch-degrading polysaccharide monooxygenases. Cell Mol Life Sci. 2016;73(14):2809-19. DOI:10.1007/s00018-016-2251-9 |
- Walton PH and Davies GJ. (2016). On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr Opin Chem Biol. 2016;31:195-207. DOI:10.1016/j.cbpa.2016.04.001 |
- Nekiunaite L, Isaksen T, Vaaje-Kolstad G, and Abou Hachem M. (2016). Fungal lytic polysaccharide monooxygenases bind starch and β-cyclodextrin similarly to amylolytic hydrolases. FEBS Lett. 2016;590(16):2737-47. DOI:10.1002/1873-3468.12293 |
- Frandsen KE, Poulsen JC, Tovborg M, Johansen KS, and Lo Leggio L. (2017). Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder. Acta Crystallogr D Struct Biol. 2017;73(Pt 1):64-76. DOI:10.1107/S2059798316019641 |
- Vaaje-Kolstad G, Forsberg Z, Loose JS, Bissaro B, and Eijsink VG. (2017). Structural diversity of lytic polysaccharide monooxygenases. Curr Opin Struct Biol. 2017;44:67-76. DOI:10.1016/j.sbi.2016.12.012 |
- Hemsworth GR, Davies GJ, and Walton PH. (2013). Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660-8. DOI:10.1016/j.sbi.2013.05.006 |
- Hemsworth GR, Henrissat B, Davies GJ, and Walton PH. (2014). Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10(2):122-6. DOI:10.1038/nchembio.1417 |