CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Polysaccharide epimerases"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | * Author: | + | * Author: [[User:Margrethe Gaardlos|Margrethe Gaardlos]] and [[User:Anne Tondervik|Anne Tondervik]] |
| − | * Responsible Curator: | + | * Responsible Curator: [[User:Finn Aachmann|Finn Aachmann]] |
---- | ---- | ||
== Introduction == | == Introduction == | ||
| Line 31: | Line 31: | ||
=== Mechanism === | === Mechanism === | ||
| − | [[Image:Mech_epimerase_lyase.png|thumb|600px|Figure 2. | + | [[Image:Mech_epimerase_lyase.png|thumb|600px|Figure 2. Three-step epimerase/lyase mechanism as proposed by Gacesa <cite> gacesa1987 </cite>. AA1, AA2 and AA3 denotes the amino acids, or potentially water molecules, responsible for each step. After neutralization of the negative charge on the D-mannuronate carboxyl group by AA1, AA2 can abstract the proton bound to C5. This results in an enolate intermediate that is resonance stabilized. A proton is subsequently donated to different groups depending on whether it is an epimerase or lyase reaction. In lyases, cleavage of the glycosidic bond forms an unsaturated residue denoted Δ and a proton is donated to the leaving group. In epimerases AA3 donates a proton to the C5 carbanion on the opposite side of the ring from the one that was abstracted, forming L-guluronate.]] |
The proposed alginate epimerase mechanism is initiated with neutralization of the negative charge of the carboxylate group by either protonation or interaction with a positively charged amino acid (Figure 2) <cite> gacesa1987 </cite>. This is followed by abstraction of H-5 by a general base residue to form an enol or enolate, followed by protonation from the opposite side of the sugar ring by a general acid residue. The conformation of the monomer flips from <sup>4</sup>''C''<sub>1</sub> to <sup>1</sup>''C''<sub>4</sub> and changes it from β-<font style="font-feature-settings: 'smcp'">d</font>-mannuronate to α-<font style="font-feature-settings: 'smcp'">l</font>-guluronate. The chemical mechanism used by alginate epimerases is believed to be similar to the mechanism of [[Polysaccharide Lyases]]. This is supported by several of the ''A. vinelandii'' enzymes having both lyase and epimerase activity <cite> ramstad1999, svanem1999, svanem2001, Holtan2006 </cite>. In the lyase mechanism, the second step is a β-elimination of the 4-''O''-glycosidic bond to form a 4-deoxy-<font style="font-feature-settings: 'smcp'">l</font>-''erythro''-hex-4-enepyranosyluronate, called Δ, at the non-reducing end. The NNHSY sequence is a common motif in both epimerases and lyases, and is believed to be important for catalysis or binding <cite> yoon2001, Ertesvag1998b, Gimmestad2003 </cite>. | The proposed alginate epimerase mechanism is initiated with neutralization of the negative charge of the carboxylate group by either protonation or interaction with a positively charged amino acid (Figure 2) <cite> gacesa1987 </cite>. This is followed by abstraction of H-5 by a general base residue to form an enol or enolate, followed by protonation from the opposite side of the sugar ring by a general acid residue. The conformation of the monomer flips from <sup>4</sup>''C''<sub>1</sub> to <sup>1</sup>''C''<sub>4</sub> and changes it from β-<font style="font-feature-settings: 'smcp'">d</font>-mannuronate to α-<font style="font-feature-settings: 'smcp'">l</font>-guluronate. The chemical mechanism used by alginate epimerases is believed to be similar to the mechanism of [[Polysaccharide Lyases]]. This is supported by several of the ''A. vinelandii'' enzymes having both lyase and epimerase activity <cite> ramstad1999, svanem1999, svanem2001, Holtan2006 </cite>. In the lyase mechanism, the second step is a β-elimination of the 4-''O''-glycosidic bond to form a 4-deoxy-<font style="font-feature-settings: 'smcp'">l</font>-''erythro''-hex-4-enepyranosyluronate, called Δ, at the non-reducing end. The NNHSY sequence is a common motif in both epimerases and lyases, and is believed to be important for catalysis or binding <cite> yoon2001, Ertesvag1998b, Gimmestad2003 </cite>. | ||
| Line 52: | Line 52: | ||
=== Catalytic residues === | === Catalytic residues === | ||
| − | [[Image:binding_actsite2.png|thumb|600px|Figure 3. Electrostatic surface potential and catalytic sites of A. AlgE4 ([{{PDBlink}}2PYH PDB ID 2PYH], <cite> Rozeboom2008 </cite>) and B. AlgG ([{{PDBlink}}4NK6 PDB ID 4NK6], <cite> wolfram2014 </cite>). The maps | + | [[Image:binding_actsite2.png|thumb|600px|Figure 3. Electrostatic surface potential and catalytic sites of A. AlgE4 ([{{PDBlink}}2PYH PDB ID 2PYH], <cite> Rozeboom2008 </cite>) and B. AlgG ([{{PDBlink}}4NK6 PDB ID 4NK6], <cite> wolfram2014 </cite>). The maps were created using the APBS electrostatics plug-in <cite> jurrus2018 </cite> in PyMOL <cite> PyMOL </cite>. The scale is from -5 to +5 kT/e, where negative potential is colored red and positive potential is colored blue. The mannuronate trimer in the crystal structure of AlgE4 is shown as ball-and-stick with carbon atoms in green and oxygen atoms in red.]] |
| − | All | + | All known alginate epimerases share a YG(F/I)DPH(D/E) motif located in the +1 subsite. In the AlgE epimerases the catalytic residues are identified as the four essential amino acids Y149, D152, and H154 of this motif, in addition to D178 that is lacking in AlgG <cite> svanem2001, nyvall2003, Douthit2005, Rozeboom2008 </cite> (See Figure 3A). The exact role of each residue in the mechanism is unclear. It has been suggested that tyrosine acts as the base in the reaction, marked AA2 in Figure 2, while the histidine is the acid, AA3 <cite> Rozeboom2008 </cite>. The two acidic residues might be important in maintaining the pK<sub>a</sub> values of amino acids in the active site, as well as orienting the catalytic base. In AlgG (Figure 3B), H319 is hypothesized to be the base, water to be the acid, and R345 to 'neutralize' the carboxyl group <cite> wolfram2014 </cite>. |
=== Role of calcium === | === Role of calcium === | ||
| Line 76: | Line 76: | ||
== Heparosan-N-sulfate-glucuronate 5-epimerase == | == Heparosan-N-sulfate-glucuronate 5-epimerase == | ||
=== Substrate specificities === | === Substrate specificities === | ||
| − | [[Image:Heparansulfate.png|thumb| | + | [[Image:Heparansulfate.png|thumb|600px|Figure 5. Hexamer illustrating the most common monomers found in heparan sulfate. GlcNAc represents α-<font style="font-feature-settings: 'smcp'">d</font>-''N''-acetyl glucosamine, GlcA is β-<font style="font-feature-settings: 'smcp'">d</font>-glucuronate, GlcNS is 2-sulfamido-α-<font style="font-feature-settings: 'smcp'">d</font>-glucosamine, IdoA(2S) is 2-''O''-sulfo-α-<font style="font-feature-settings: 'smcp'">l</font>-iduronate, GlcNS(6S) is 2-sulfamido-α-<font style="font-feature-settings: 'smcp'">d</font>-glucosamine-6-''O''-sulfate and IdoA is α-<font style="font-feature-settings: 'smcp'">l</font>-iduronate. Glucosamine sulfated at 3-O (GlcNS(3S,6S)) and glucosamine with a free amine group (GlcN) also exist, but these are rarer and are not shown in the structure.]] |
| − | + | Heparin/heparan epimerase catalyzes epimerization of β-<font style="font-feature-settings: 'smcp'">d</font>-glucuronic acid (GlcA) to α-<font style="font-feature-settings: 'smcp'">l</font>-iduronic acid (IdoA) at the polymer level in the glycosaminoglycan heparan sulfate <cite>hook1974</cite>. Heparan sulfate consists of long chains of a repeating disaccharide motif, 1,4-linked alternating monomers of GlcA or IdoA and α-<font style="font-feature-settings: 'smcp'">d</font>-glucosamine (GlcN). GlcN monomers can be ''N''-acetylated or ''N''-sulfated, and all three sugar units can be ''O''-sulfated. While GlcA adopts a <sup>4</sup>''C''<sub>1</sub> chair conformation, IdoA exists in an equilibrium between <sup>1</sup>''C''<sub>4</sub> and the skew-boat conformation <sup>2</sup>''S''<sub>0</sub> <cite>valla2001</cite>. Domains in heparan sulfate have various compositions of monomers and modifications, and long domains with high amounts of ''N''-sulfated disaccharide units are called heparin <cite>gallagher1985</cite>. Figure 5 shows a hexamer with common heparan sulfate monomers. | |
| − | Heparan sulfate is found in all animal cells as proteoglycans. Due to its negative charge and diverse sequence motifs, the polysaccharide | + | Heparan sulfate is found in all animal cells as proteoglycans. Due to its negative charge and diverse sequence motifs, the polysaccharide interacts with many different proteins and affect most cellular processes mainly through electrostatic interactions. Heparin is stored in secretory granules in mast cells and released when tissue is injured. When it binds to antithrombin it aids in inhibiting blood coagulation. Heparin is produced industrially as an anticoagulant; its roles in tissue injury and repair in cells are diverse and complex <cite>valla2001, bishop2007</cite>. |
| − | The activity of the heparin/heparan epimerase requires that the residue to be epimerised has a deacetylated and ''N''-sulfated glucosamine linked to its C-4 (subsite -1). ''O''- | + | The activity of the heparin/heparan epimerase requires that the GlcA/IdoA residue to be epimerised has a deacetylated and ''N''-sulfated glucosamine linked to its C-4 (subsite -1). ''O''-Sulfation of the residue to be epimerized, or of neighboring residues, inhibits epimerization. The residue linked to C-1 of the sugar unit in the active site can be ''N''-sulfated or ''N''-acetylated, but ''N''-sulfation yields a better substrate. An octasaccharide is the minimal substrate size <cite>jacobsson1984, hagner2000</cite>. |
| − | The enzymatic reaction is reversible when the substrate has a glucosamine or sulfated glucosamine three residues away from the epimerization site towards the nonreducing end, or if this site is unoccupied. | + | The enzymatic reaction is reversible when the substrate has a glucosamine or sulfated glucosamine three residues away from the epimerization site towards the nonreducing end, or if this site is unoccupied. Conversely, if the substrate has an acetylated glucosamine in the same site the enzyme irreversibly introduces <font style="font-feature-settings: 'smcp'">l</font>-iduronic acid residues to the heparin chains. Because IdoA residues are substrates for sulfation reaction in vivo, and the resulting ''O''-sulfated IdoA is not a substrate, epimerization is effectively irreversible in vivo <cite>hagner2004, li2010, sheng2012</cite>. |
| − | An enzyme with a | + | An enzyme with a different substrate specificity has been identified from the giant African snail ''Achatina fulica'' <cite>mochizuki2015</cite>. The glycosaminoglycan acharan sulfate contains repeating units of ''N''-acetylated glucosamine and ''O''-sulfated IdoA. The epimerase creating IdoA in this polymer is named heparosan-glucuronate 5-epimerase since it is active on heparosan, as opposed to the heparosan-N-sulfate-glucuronate 5-epimerase that requires a deacetylated and sulfated substrate. |
=== Catalytic reaction and mechanism === | === Catalytic reaction and mechanism === | ||
| − | The catalytic reaction is thought to follow a similar mechanism to the mannuronan C5-epimerases described [[#Mechanism|above]], and also to that of heparin and dermatan sulfate lyases <cite>Han2009,lunin2004</cite>. A tyrosine | + | The catalytic reaction is thought to follow a similar mechanism to the mannuronan C5-epimerases described [[#Mechanism|above]], and also to that of heparin and dermatan sulfate lyases <cite>Han2009,lunin2004</cite>. A tyrosine acts as the proton acceptor and a glutamate as the proton donor. Based on structural analysis of substrate and product complexes the reaction is believed to proceed through a neutral enol intermediate, with ring distortion to facilitate epimerization <cite>debarnot2019</cite>. Originally the intermediate was proposed to be an α-carbanion/enolate <cite>prihar1980</cite>, as for the other polysaccharide epimerases <cite>qin2015</cite>. Two other tyrosines are essential for activity, and they are hypothesized to be part of proton relay pathways between the active site and the solvent <cite>qin2015, debarnot2019</cite>. Protonation of the enol is the rate-limiting step <cite>hagner2000b, debarnot2019</cite>. Heparin/heparan epimerases do not require divalent cations for activity, unlike the alginate and dermatan sulfate epimerases <cite>malmstrom1980</cite>. However, one calcium ion is bound to each subunit in the crystal structure of human heparin/heparan epimerase <cite>debarnot2019</cite>. In the crystal structure of zebrafish heparin/heparan epimerase, a water molecule is bound in the same position <cite>qin2015</cite>. |
=== Three-dimensional structures === | === Three-dimensional structures === | ||
| − | [[Image:hglce.jpg|thumb| | + | [[Image:hglce.jpg|thumb|600px|Figure 6. Cartoon representation of the dimeric assembly of human glucuronyl C5-epimerase ([{{PDBlink}}6HZZ PDB ID 6HZZ]) <cite>debarnot2019</cite>. The N-terminal β-hairpin domain is colored in purple, the β-sandwich domain is colored in red and the (α/α)<sub>4</sub>-barrel domain is colored in green. One calcium ion is coordinated in the sandwich domain of each subunit and shown as an orange sphere. Three ''N''-glycans bound to each subunit are shown in sticks and spheres.]] |
| − | + | The first crystal structure was reported in 2015, <font style="font-feature-settings: 'smcp'">d</font>-glucuronyl C5-epimerase from zebrafish ([{{PDBlink}}4PW2 PDB ID 4PW2]) <cite>qin2015</cite>. The protein crystallizes as a homodimer and has three domains: an α-helical transmembrane region, a β-barrel domain, and a flexible N-terminal loop. An enzyme complex with an inhibiting heparin hexamer was also reported that defined the active site cleft ([{{PDBlink}}4PXQ PDB ID 4PXQ]). | |
| − | The | + | The structure of human <font style="font-feature-settings: 'smcp'">d</font>-glucuronyl C5-epimerase has been reported ([{{PDBlink}}6HZZ PDB ID 6HZZ]) <cite>debarnot2019</cite>. Like the zebrafish enzyme, it is a dimer where each subunit has three domains. The N-terminal domain consists of two antiparallel β-hairpins connected by a helix. This is followed by a β-sandwich domain, and a C-terminal (α/α)<sub>4</sub>-barrel domain containing the active site. The dimer is probably the active form of the enzyme. Figure 6 shows the dimeric structure of human glucuronyl C5-epimerase. Structures of an inactive mutant bound to a substrate and a product were also deposited (PDB IDs [{{PDBlink}}6I01 6I01] and [{{PDBlink}}6I02 6I02]) <cite>debarnot2019</cite>. |
== Dermatan sulfate C5-epimerase == | == Dermatan sulfate C5-epimerase == | ||
=== Substrate specificities === | === Substrate specificities === | ||
| − | [[Image:Dermatansulfate.png|thumb|600px|Figure 7. | + | [[Image:Dermatansulfate.png|thumb|600px|Figure 7. Heptamer motif of a dermatan sulfate chain showing the possible monomers. GalNAc(4S,6S) represents β-<font style="font-feature-settings: 'smcp'">d</font>-''N''-acetyl galactosamine 4,6-''O''-sulfate, GlcA is β-<font style="font-feature-settings: 'smcp'">d</font>-glucuronate, GalNAc is β-<font style="font-feature-settings: 'smcp'">d</font>-''N''-acetyl galactosamine, IdoA is α-<font style="font-feature-settings: 'smcp'">l</font>-iduronate, GalNAc(4S) is β-<font style="font-feature-settings: 'smcp'">d</font>-''N''-acetyl galactosamine 4-''O''-sulfate, IdoA(2S) is 2-''O''-sulfo-α-<font style="font-feature-settings: 'smcp'">l</font>-iduronate and GalNAc(6S) is β-<font style="font-feature-settings: 'smcp'">d</font>-''N''-acetyl galactosamine 6-''O''-sulfate.]] |
| − | Dermatan sulfate epimerases catalyze the epimerization of β-<font style="font-feature-settings: 'smcp'">d</font>-glucuronic acid (GlcA) to α-<font style="font-feature-settings: 'smcp'">l</font>-iduronic acid (IdoA) in polymers of glycosaminoglycans <cite>malmstrom1982, maccarana2006, pacheco2009b</cite>, an activity similar to heparin epimerase. The repeating disaccharide unit of dermatan sulfate is 1- | + | Dermatan sulfate epimerases catalyze the epimerization of β-<font style="font-feature-settings: 'smcp'">d</font>-glucuronic acid (GlcA) to α-<font style="font-feature-settings: 'smcp'">l</font>-iduronic acid (IdoA) in polymers of glycosaminoglycans <cite>malmstrom1982, maccarana2006, pacheco2009b</cite>, an activity similar to heparin epimerase. The repeating disaccharide unit of dermatan sulfate is 1,3-linked GlcA or IdoA and 1,4-linked β-<font style="font-feature-settings: 'smcp'">d</font>-''N''-acetyl galactosamine (GalNAc). Sulfation can occur at C-2 of iduronic acid and C-4 and/or C-6 of galactosamine. Figure 7 illustrates the seven possible monomers after sulfation and epimerization, three for the hexuronates and four for galactosamine. Dermatan sulfate can have various lengths, degrees of epimerization, and sulfation patterns, and can bind to a variety of proteins <cite>trowbridge2002</cite>. The unepimerized polymer is called chondroitin sulfate. Although dermatan sulfate was earlier known as chondroitin B it is no longer classified as a chondroitin sulfate <cite>NLM2020</cite>. Chondroitin/dermatan sulfate exists as proteoglycans in extracellular matrixes in mammalian tissues, especially skin. The polysaccharide is involved in many different cell processes due to its sequence variability and diverse protein partners <cite>trowbridge2002</cite>. |
| − | Dermatan sulfate consists of block structures of (IdoA-GalNAc)n, (GlcA-GalNAc)n, and hybrid domains containing both uronic acids. Two epimerases are known: dermatan sulfate epimerases 1 and 2, which have slightly different product patterns and which could be important in regulating domain formation. They are found in a variety of animals, including humans <cite>pacheco2009b</cite>. | + | Dermatan sulfate consists of block structures of (IdoA-GalNAc)<sub>n</sub>, (GlcA-GalNAc)<sub>n</sub>, and hybrid domains containing both uronic acids. Two epimerases are known: dermatan sulfate epimerases 1 and 2, which have slightly different product patterns and which could be important in regulating dermatan sulfate domain formation. They are found in a variety of animals, including humans <cite>pacheco2009b</cite>. |
Dermatan is a better substrate for the epimerases than chondroitin, and the epimerases are inactive on sulfated substrates <cite>malmstrom1984</cite>. However, the co-incubation of epimerases and sulfotransferases synergistically promotes the formation of iduronic acid. The epimerases physically interact with the sulfotransferase and processively form long iduronic acid-containing domains <cite>malmstrom1975, tykesson2018</cite>. The optimal minimal substrate is an octamer <cite>tykesson2016</cite>. | Dermatan is a better substrate for the epimerases than chondroitin, and the epimerases are inactive on sulfated substrates <cite>malmstrom1984</cite>. However, the co-incubation of epimerases and sulfotransferases synergistically promotes the formation of iduronic acid. The epimerases physically interact with the sulfotransferase and processively form long iduronic acid-containing domains <cite>malmstrom1975, tykesson2018</cite>. The optimal minimal substrate is an octamer <cite>tykesson2016</cite>. | ||
Latest revision as of 13:16, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: Margrethe Gaardlos and Anne Tondervik
- Responsible Curator: Finn Aachmann
Introduction
Epimerases and racemases catalyse the inversion of stereochemistry in substrate molecules. Epimerases invert the stereochemistry of only one stereogenic center in a molecule that has at least two, whereas racemases catalyze the racemization of pure enantiomers, usually through the inversion of a stereogenic center in a molecule that possesses just one. These enzymes allow organisms to utilize several different stereoisomers of the same compounds. Whereas most enzymes catalyze reactions where reactants and products have only one stereochemistry, epimerases and racemases must be able to accommodate two different stereoisomers in their active site. Many epimerases and racemases act by breaking an acidic C-H bond next to a carbonyl functional group to give an enol or enolate, and then reform it with the new stereochemistry [1], while others break a C-H bond of an alcohol, acting through an oxidation/reduction sequence.
van Overtveldt et al. classified 39 known carbohydrate epimerase specificities into 14 different families based on sequence alignments [2]. Five different epimerization mechanisms were described: I) deprotonation/reprotonation, II) transient keto intermediate, III) carbon-carbon bond cleavage, IV) nucleotide elimination, and V) mutarotation. Members of a family share mechanism and conserved catalytic residues. Only two of the 14 families are active on polysaccharides: CEP7 containing mannuronan C5-epimerases (alginate epimerases) and CEP8 containing heparosan-N-sulfate-glucuronate 5-epimerase (heparosan epimerase). Together with the unclassified dermatan sulfate C5-epimerase (chondroitin epimerase), these enzymes perform a similar 'polymer-level' C5-epimerization on polysaccharides consisting of hexuronic acids. Alginates and the glycosaminoglycans heparin/heparan and dermatan sulfate are block copolymers of the two monomers created by these epimerases. In alginates, the epimerization is from d-mannuronic acid to l-guluronic acid, while in the glycosaminoglycans it is from d-glucuronic acid to l-iduronic acid [3, 4, 5, 6].
Alginate and glucosaminoglycan epimerases utilize a deprotonation/reprotonation mechanism where neutralization of the sugar carboxyl group is followed by proton abstraction from one side of the sugar ring, creating a resonance stabilized enol or enolate intermediate. The pKa value of the hydrogen is lowered due to this resonance stabilization [1, 6]. For the mannuronan C5-epimerase, the intermediate is thought to be an enolate anion, while for the heparin epimerase it could be a neutral enol [7]. For dermatan epimerases, it is proposed that deprotonation at C5 leads to elimination of the sugar attached at C4, with cleavage of the bond to the glycosidic oxygen, affording a 4,5-unsaturated intermediate that undergoes an addition reaction to reform the glycoside but with the proton added from the opposite side to give the epimer [8]; this elimination/addition mechanism has also been proposed as an alternative for alginate epimerases [9]. The mechanism is probably similar to that of Polysaccharide Lyases, which undergo a β-elimination to afford a 4,5-unsaturated hexenuronic acid product [10].
Despite performing similar reactions, the three hexuronyl C5-epimerase families share almost no sequence or structural similarity [2, 11].
Mannuronan C5-epimerases
Substrate specificity
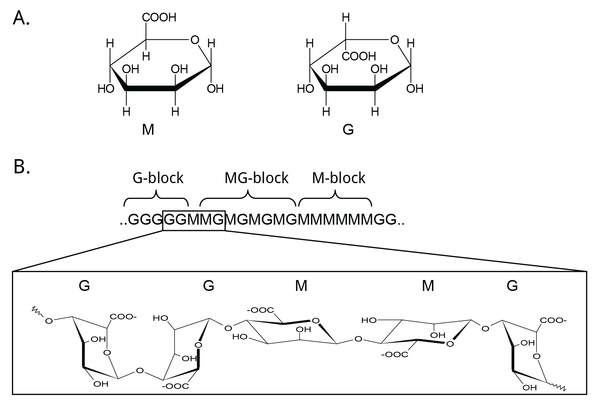
Mannuronan C5-epimerases are a group of enzymes that catalyze epimerization at the polymer-level of β-d-mannuronic acid residues (hereafter denoted M) into α-l-guluronic acid residues (hereafter denoted G) in alginate [3, 12, 13]. Alginate is an anionic polysaccharide made by brown seaweeds, some species of red algae, and bacteria of the gram-negative genera Pseudomonas and Azotobacter [14, 15, 16, 17, 18]. The functions of alginate in different organisms relate to structure, protection, and surface adhesion [19, 20, 21, 22]. Alginate is a copolymer of the two 1,4-linked epimers [23, 24, 25], and by changing the relative amounts of the M and G monomers the epimerases fine-tune the properties of alginate [26]. Initially, alginate is made as a homopolymer of M in the cell. Epimerases then convert some of the M-residues in the polymer into G-residues [13, 27, 28]. This epimerization is not random; it creates block structures of M, G, or alternating MG [29, 30], see Figure 1. Alginate residues that are oxidized or acetylated are not substrates for the epimerases, and acetylation of alginate could be a way to control epimerization [31, 32].
Mannuronan C5-epimerases exist both in algae and in bacteria [3, 33]. Gene analyses propose as many as 31 different genes encoding putative mannuronan C-5 epimerases in the brown algae Ectocarpus [34]. However, the algal epimerases are difficult to express and as a result the bacterial enzymes have been studied most extensively [34, 35]. Two categories of bacterial mannuronan C5-epimerases have been described: the periplasmic AlgG and the extracellular and calcium-dependent AlgE. AlgG creates single G residues in stretches of mannuronan, while the AlgE enzymes are processive and create MG-blocks and G-blocks. Pseudomonas spp. are only known to produce AlgG [28, 36, 37], while A. vinelandii contains seven active AlgE enzymes, AlgE1-7, in addition to AlgG [38, 39, 40, 41]. Recently, three AlgE enzymes were characterized in A. chroococcum and denoted AcAlgE1-3 [42]. A mutant strain of P. fluorescens without the algG gene creates pure mannuronan [43]. This strain can be used to produce an unepimerized substrate, which is useful for the study of the epimerization reaction. Methods for studying this are discussed below.
Product profiles
The assorted epimerases of A. vinelandii give slightly different product profiles that makes it possible for the bacterium to tailor its alginate structure to fulfill different functions [39, 44]. This is done in three different ways. Firstly, some AlgE enzymes can only create MG-blocks, while others also create G-blocks. Secondly, different epimerases create block stretches of different lengths. Lastly, one of the seven A. vinelandii AlgE epimerases (AlgE7) has a dual epimerase/lyase activity and thus modifies the polymer length [41]. Two of the three AlgE enzymes found in A. chroococcum are homologous to AlgE7 and display the same bifunctional activity [42, 45]. Weak lyase activity has been observed in other AlgEs, although it is not certain whether this serves a function or if it is the result of failed epimerization reactions [46, 47].
Catalytic reaction
The extracellular A. vinelandii AlgE enzymes consist of different combinations of a catalytic module, the A-module, and a smaller R-module that is thought to modify binding [44, 48]. The enzymes' direction of movement along the substrate is unknown, but there are indications that they move along their polymeric substrate from the non-reducing to the reducing end [47, 49]. The epimerases show various degrees of processivity [47, 50, 51, 52], with AlgE4 catalyzing around 10-12 epimerizations before disassociating from the substrate [49, 53]. Epimerases are thought to only epimerize every other residue in one binding event, which means that the G-block formers will need to bind the MG-product of the first reaction again to form G-blocks [47, 49]. The reason why some epimerases can not form G-blocks may be related to their interactions with poly-MG sequences [53].
Mechanism
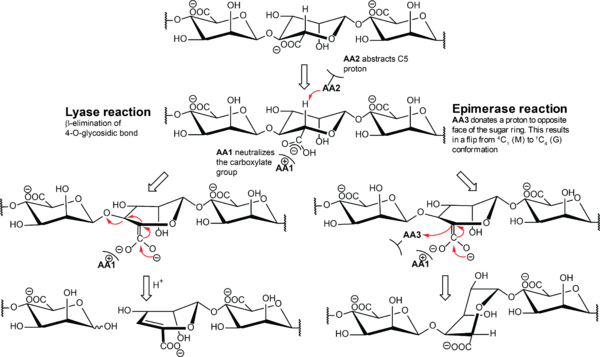
The proposed alginate epimerase mechanism is initiated with neutralization of the negative charge of the carboxylate group by either protonation or interaction with a positively charged amino acid (Figure 2) [10]. This is followed by abstraction of H-5 by a general base residue to form an enol or enolate, followed by protonation from the opposite side of the sugar ring by a general acid residue. The conformation of the monomer flips from 4C1 to 1C4 and changes it from β-d-mannuronate to α-l-guluronate. The chemical mechanism used by alginate epimerases is believed to be similar to the mechanism of Polysaccharide Lyases. This is supported by several of the A. vinelandii enzymes having both lyase and epimerase activity [41, 46, 47, 54]. In the lyase mechanism, the second step is a β-elimination of the 4-O-glycosidic bond to form a 4-deoxy-l-erythro-hex-4-enepyranosyluronate, called Δ, at the non-reducing end. The NNHSY sequence is a common motif in both epimerases and lyases, and is believed to be important for catalysis or binding [43, 55, 56].
Methods to study the reaction
Several methods have been used either to measure catalytic rates or to characterize the epimerized product in terms of relative amounts of M, G, and block compositions.
The Dische carbazole reaction [57] can be used to measure both initial activity and endpoint conversion [13, 58, 59]. In this method, an increase in color intensity from mannuronic to guluronic acid is used to quantify the degree of epimerization.
Epimerization activity on 5-3H-alginate can be measured by observing tritium released into the solvent [60, 61]. This method has increased accuracy compared to the carbazole method is better suited for determination of kinetic parameters. Notably, because the substrate changes as epimerization occurs, classical Michaelis-Menten kinetics do not apply; instead, apparent values for Vmax and kcat are obtained. For AlgE4 they were determined to be 14.8 μmol min-1 mg-1 protein and 14 s-1, respectively [50].
A fast and sensitive method that does not require tritiated alginate involves the direct detection of the unsaturated product of the alginate lyase reaction, Δ, which absorbs at 230 nm [62]. This can be used to measure lyase activity directly [63], but it can also be used to measure epimerization in a coupled assay. This is done by treating epimerized alginate with an alginate lyase, e.g., AlyA from Klebsiella pneumoniae, which specifically cleaves at G-M and G-G linkages [64]. Formation of Δ, monitored by measuring absorbance at 230 nm, is then assumed to be directly proportional to the amount of G produced by the epimerase.
Block composition can be assessed by acid hydrolysis of alginate [3, 29, 30]. Alternating blocks are readily hydrolysed and appear in the soluble fraction, while homopolymeric blocks are less reactive and remain insoluble. Subsequently, dissolution of the insoluble part followed by acid precipitation of G-blocks, allows the relative amounts of the three block structures to be roughly estimated. A more precise method uses either 13C-NMR [65, 66] or the more sensitive 1H-NMR [67, 68] spectroscopy to calculate block composition of alginate. This is done by measuring relative amounts of monomer dyads and triads.
Both 13C- and 1H-NMR spectroscopy have been used to monitor rates of epimerization, allowing measurement of kinetics and observation of the mode of action [54, 69, 70, 71].
Circular dichroism can distinguish between M and G [72, 73], and can be used to measure epimerase activity [74].
To determine block length and distribution, lyases with four different specificities (cutting the alginate chain at either M-M, G-G, M, or G) can be used [75]. Size exclusion chromatography (SEC) is used to separate the products and the resulting block fractions are analyzed with high-pH anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) [76, 77], NMR spectroscopy, or SEC with multiple angle light scattering (SEC-MALS) [78].
Catalytic residues
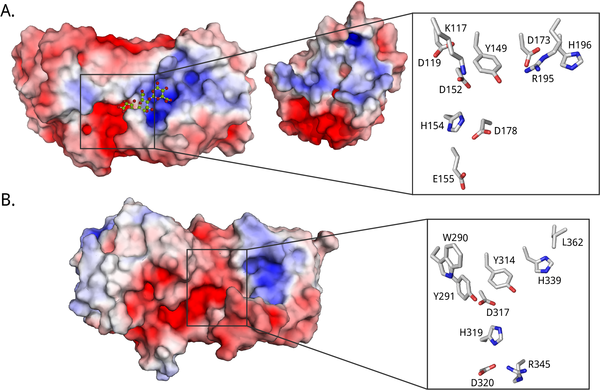
All known alginate epimerases share a YG(F/I)DPH(D/E) motif located in the +1 subsite. In the AlgE epimerases the catalytic residues are identified as the four essential amino acids Y149, D152, and H154 of this motif, in addition to D178 that is lacking in AlgG [35, 54, 79, 83] (See Figure 3A). The exact role of each residue in the mechanism is unclear. It has been suggested that tyrosine acts as the base in the reaction, marked AA2 in Figure 2, while the histidine is the acid, AA3 [79]. The two acidic residues might be important in maintaining the pKa values of amino acids in the active site, as well as orienting the catalytic base. In AlgG (Figure 3B), H319 is hypothesized to be the base, water to be the acid, and R345 to 'neutralize' the carboxyl group [80].
Role of calcium
The seven secreted epimerases of A. vinelandii, AlgE1-AlgE7, are calcium-dependent [3, 44, 84]. Activity increases with increasing calcium concentration up to around 5 mM calcium, with the optimum depending on the enzyme [13, 46, 50, 54, 85]. At the same time, increased calcium concentration gives a decrease in end-point conversion due to gelation of the product [13]. It is unknown whether calcium is important for the mechanism itself or if its role is to stabilize the structure (see three-dimensional structures below). Early studies indicated a role in enzyme stability, as the temperature stability increased at higher calcium concentrations [13]. In the 3D crystal structure of AlgE4's A-module, a calcium ion is bound in a hairpin loop away from the active site [79] (PDB ID 2PYH). Without the calcium ion, the loop would likely change conformation, which could destabilize the active fold of the enzyme. Calcium may also be involved in neutralizing the negative charge of the carboxylate anion, which is observed in the structurally similar alginate lyases from the polysaccharide lyase family 6 [86, 87, 88, 89]. The periplasmic AlgG epimerases do not require calcium for activity [28, 40].
Substrate binding
Alginate is a polyelectrolyte, and substrate binding probably occurs mainly through electrostatic interactions. The A. vinelandii epimerases are modular, and together the modules form an extended binding groove lined by charged residues [48, 90] (see Figure 3 for electrostatic surface potential maps). Assuming a monomer length of about 0.5 nm [53] and a binding groove length of about 5 nm [79], the catalytically active A-module can accommodate around 10 sugar residues. The R-module seems to be able to bind around 5 alginate monomers [48]. The epimerases probably require different substrate lengths to be able to initiate epimerization. For instance, AlgE4 seems to need at least a hexamer [49], while AlgE6 and AlgE1 need eight to ten monomers [47].
Different epimerases create different block lengths: the larger epimerase AlgE1 appears to create longer blocks than AlgE6, which in turn seems to create longer blocks than AlgE4 [47, 49]. This could be due to differences in processivity. Since the alginate epimerases are processive, high affinity substrate binding may decrease their activity. The R-modules may have a role in processivity: even though the R-module from AlgE4 increases its binding compared to the A-module alone, R-modules from AlgE6 cannot individually tobind alginate [48]. The R-modules probably modulate binding and processivity in complex ways, not solely by increasing the binding affinity [44, 48, 53, 70, 90, 91].
Residue 307 has been highlighted as important for epimerase substrate specificity. It is located in a large loop in the A-module of the AlgE enzymes. In G-block formers, this residue is tyrosine, whereas in MG-block formers it is a phenylalanine [70]. Mutations of this residue can change the epimerization pattern [71]. Because this residue is located relatively distant from the active site it it suggests that substrate binding, and not the catalytically active residues, that gives rise to different product profiles for different alginate epimerases.
Three-dimensional structures
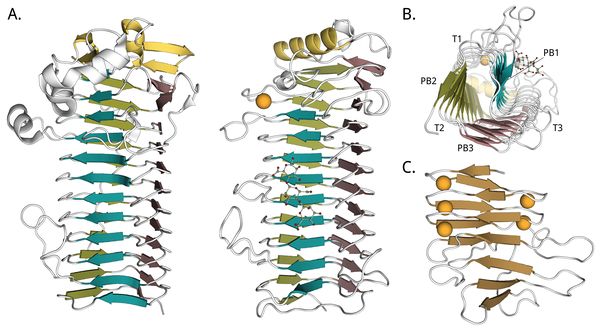
The first 3D crystal structure of a mannuronan C5-epimerase was that of the AlgE4 A-module from A. vinelandii, which was solved at 2.1 Å resolution [79]. It shows a right-handed parallel β-helix fold with an N-terminal α-helix cap and an extended binding groove, see Figure 4B. Protruding from the binding groove are three flexible loops, slightly enclosing the binding surface. One of the two molecules of the asymmetric unit has a mannuronan trimer bound in its binding groove. A calcium ion is coordinated in proximity to the active site, at the N-terminal end. The second 3D crystal structure reported was of the P. aeruginosa non-modular AlgG epimerase, at 2.1 Å resolution [80] (Figure 4A). It is structurally similar to AlgE4's A-module. In 2016 the A-module of AlgE6 was deposited in the PDB (PDB ID 5LW3) at 1.19 Å resolution, and it is almost identical to AlgE4's A-module. These two A-modules also share the highest sequence homology of the AlgE A-modules [41]. The three structures are all around 70 Å long. R-modules of AlgE4 [90] (PDB ID 2AGM) and AlgE6 [48] (PDB IDs 2ML1, 2ML2, and 2ML3) from A. vinelandii were solved by NMR. The R-modules have an overall ellipsoid or spherical shape, continuing the parallel β-sheets of the A-module with a parallel β-roll fold (See Figure 4C). A common feature of this fold is a repeated nonapeptide motif LXGGAGXDXn, a circular permutation of the motif GGXGXDX(L/I/F)X first found in RTX (repeats in toxins) toxins from Gram-negative bacteria [93, 94]. The motifs stabilize the fold by binding calcium ions tightly [95], and the R-modules of AlgE contains four to seven of them [26, 90]. The core part of the R-modules is around 40 Å long. At the C-terminal of the last R-module of each enzyme is an unstructured region of about 20 amino acids. This is thought to function as a secretion signal for a transporter that secretes the enzymes out of the cell [38, 90, 96].
No structures have been reported for a complete multimodular epimerase. From low resolution SAXS-measurements of AlgE4 and AlgE6, the enzymes appear elongated, with the R-modules extending the binding grooves of the A-modules [48]. Some flexibility between the modules is observed, and NMR-studies indicate a flexible linker between the A- and the R-modules. Radii of gyration are calculated to be 31 Å for AlgE4 and from 52-55 Å for AlgE6. Maximum distances are around 100 Å for AlgE4 and around 180 Å for AlgE6 [48].
The parallel β-helix fold found in the active modules of the epimerases was first encountered in another enzyme active on a polyanionic substrate, namely the pectate lyase C of Erwinia chrysanthemi [97]. It is also called a β-solenoid-type fold as it consists of repeated β strands, and it is a type L β-solenoid [98]. In the epimerases, the β-solenoid is found together with an N-terminal α-helix cap that shields the hydrophobic interior of the β-helix from the solvent. This combination is found in several other enzyme families of glycoside hydrolases (families GH28, GH49, GH55, GH82, and GH87 [99]) and of polysaccharide lyases (families PL1, PL3, PL6, and PL9 [100]): alginate lyases [88], dextranases [101], rhamnogalacturonases [102], polygalacturonase [103], β-1,3-glucanases [104], ι-carrageenases [105], endorhamnosidases [106], pectate and pectin lyases [97, 107, 108, 109] and chondroitin B (dermatan sulfate) lyases [86]. Since these enzymes have a high degree of structural similarity, it is thought that at least some of them diverged from a common ancestor [110], although convergent evolution is not unlikely [98].
Heparosan-N-sulfate-glucuronate 5-epimerase
Substrate specificities
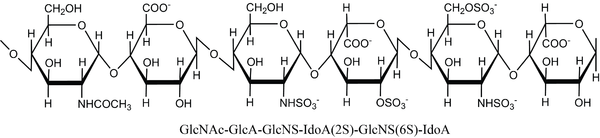
Heparin/heparan epimerase catalyzes epimerization of β-d-glucuronic acid (GlcA) to α-l-iduronic acid (IdoA) at the polymer level in the glycosaminoglycan heparan sulfate [111]. Heparan sulfate consists of long chains of a repeating disaccharide motif, 1,4-linked alternating monomers of GlcA or IdoA and α-d-glucosamine (GlcN). GlcN monomers can be N-acetylated or N-sulfated, and all three sugar units can be O-sulfated. While GlcA adopts a 4C1 chair conformation, IdoA exists in an equilibrium between 1C4 and the skew-boat conformation 2S0 [6]. Domains in heparan sulfate have various compositions of monomers and modifications, and long domains with high amounts of N-sulfated disaccharide units are called heparin [112]. Figure 5 shows a hexamer with common heparan sulfate monomers.
Heparan sulfate is found in all animal cells as proteoglycans. Due to its negative charge and diverse sequence motifs, the polysaccharide interacts with many different proteins and affect most cellular processes mainly through electrostatic interactions. Heparin is stored in secretory granules in mast cells and released when tissue is injured. When it binds to antithrombin it aids in inhibiting blood coagulation. Heparin is produced industrially as an anticoagulant; its roles in tissue injury and repair in cells are diverse and complex [6, 113].
The activity of the heparin/heparan epimerase requires that the GlcA/IdoA residue to be epimerised has a deacetylated and N-sulfated glucosamine linked to its C-4 (subsite -1). O-Sulfation of the residue to be epimerized, or of neighboring residues, inhibits epimerization. The residue linked to C-1 of the sugar unit in the active site can be N-sulfated or N-acetylated, but N-sulfation yields a better substrate. An octasaccharide is the minimal substrate size [114, 115].
The enzymatic reaction is reversible when the substrate has a glucosamine or sulfated glucosamine three residues away from the epimerization site towards the nonreducing end, or if this site is unoccupied. Conversely, if the substrate has an acetylated glucosamine in the same site the enzyme irreversibly introduces l-iduronic acid residues to the heparin chains. Because IdoA residues are substrates for sulfation reaction in vivo, and the resulting O-sulfated IdoA is not a substrate, epimerization is effectively irreversible in vivo [116, 117, 118].
An enzyme with a different substrate specificity has been identified from the giant African snail Achatina fulica [119]. The glycosaminoglycan acharan sulfate contains repeating units of N-acetylated glucosamine and O-sulfated IdoA. The epimerase creating IdoA in this polymer is named heparosan-glucuronate 5-epimerase since it is active on heparosan, as opposed to the heparosan-N-sulfate-glucuronate 5-epimerase that requires a deacetylated and sulfated substrate.
Catalytic reaction and mechanism
The catalytic reaction is thought to follow a similar mechanism to the mannuronan C5-epimerases described above, and also to that of heparin and dermatan sulfate lyases [120, 121]. A tyrosine acts as the proton acceptor and a glutamate as the proton donor. Based on structural analysis of substrate and product complexes the reaction is believed to proceed through a neutral enol intermediate, with ring distortion to facilitate epimerization [7]. Originally the intermediate was proposed to be an α-carbanion/enolate [122], as for the other polysaccharide epimerases [11]. Two other tyrosines are essential for activity, and they are hypothesized to be part of proton relay pathways between the active site and the solvent [7, 11]. Protonation of the enol is the rate-limiting step [7, 123]. Heparin/heparan epimerases do not require divalent cations for activity, unlike the alginate and dermatan sulfate epimerases [124]. However, one calcium ion is bound to each subunit in the crystal structure of human heparin/heparan epimerase [7]. In the crystal structure of zebrafish heparin/heparan epimerase, a water molecule is bound in the same position [11].
Three-dimensional structures
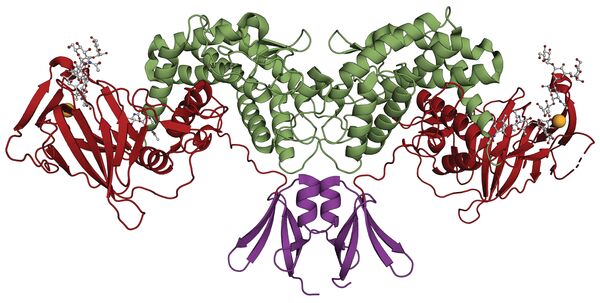
The first crystal structure was reported in 2015, d-glucuronyl C5-epimerase from zebrafish (PDB ID 4PW2) [11]. The protein crystallizes as a homodimer and has three domains: an α-helical transmembrane region, a β-barrel domain, and a flexible N-terminal loop. An enzyme complex with an inhibiting heparin hexamer was also reported that defined the active site cleft (PDB ID 4PXQ).
The structure of human d-glucuronyl C5-epimerase has been reported (PDB ID 6HZZ) [7]. Like the zebrafish enzyme, it is a dimer where each subunit has three domains. The N-terminal domain consists of two antiparallel β-hairpins connected by a helix. This is followed by a β-sandwich domain, and a C-terminal (α/α)4-barrel domain containing the active site. The dimer is probably the active form of the enzyme. Figure 6 shows the dimeric structure of human glucuronyl C5-epimerase. Structures of an inactive mutant bound to a substrate and a product were also deposited (PDB IDs 6I01 and 6I02) [7].
Dermatan sulfate C5-epimerase
Substrate specificities
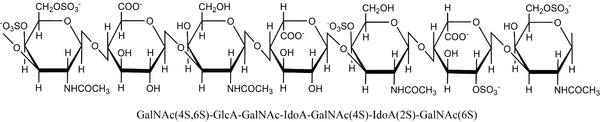
Dermatan sulfate epimerases catalyze the epimerization of β-d-glucuronic acid (GlcA) to α-l-iduronic acid (IdoA) in polymers of glycosaminoglycans [125, 126, 127], an activity similar to heparin epimerase. The repeating disaccharide unit of dermatan sulfate is 1,3-linked GlcA or IdoA and 1,4-linked β-d-N-acetyl galactosamine (GalNAc). Sulfation can occur at C-2 of iduronic acid and C-4 and/or C-6 of galactosamine. Figure 7 illustrates the seven possible monomers after sulfation and epimerization, three for the hexuronates and four for galactosamine. Dermatan sulfate can have various lengths, degrees of epimerization, and sulfation patterns, and can bind to a variety of proteins [128]. The unepimerized polymer is called chondroitin sulfate. Although dermatan sulfate was earlier known as chondroitin B it is no longer classified as a chondroitin sulfate [129]. Chondroitin/dermatan sulfate exists as proteoglycans in extracellular matrixes in mammalian tissues, especially skin. The polysaccharide is involved in many different cell processes due to its sequence variability and diverse protein partners [128].
Dermatan sulfate consists of block structures of (IdoA-GalNAc)n, (GlcA-GalNAc)n, and hybrid domains containing both uronic acids. Two epimerases are known: dermatan sulfate epimerases 1 and 2, which have slightly different product patterns and which could be important in regulating dermatan sulfate domain formation. They are found in a variety of animals, including humans [127].
Dermatan is a better substrate for the epimerases than chondroitin, and the epimerases are inactive on sulfated substrates [130]. However, the co-incubation of epimerases and sulfotransferases synergistically promotes the formation of iduronic acid. The epimerases physically interact with the sulfotransferase and processively form long iduronic acid-containing domains [5, 131]. The optimal minimal substrate is an octamer [132].
Catalytic reaction and mechanism
A mechanism is suggested where a proton is abstracted from one side of glucuronic acid by a histidine, the glycosidic bond is cleaved by the involvement of a tyrosine and another histidine attaching a proton to the other side of the ring. This will involve a 4,5-unsaturated intermediate, similar to the end product of polysaccharide lyases. The glycosidic bond would be formed again by the tyrosinate ion or a second general acid [8]. The two histidines could switch roles as acid and base, giving rise to the reversibility seen in vitro [133]. The enzyme requires divalent cations, preferably Mn2+, and N-glycosylation for activity [8, 125]. It processively epimerizes four to five monomers from the reducing towards the non-reducing end, with decreased binding affinity for each processive step [132]. In vivo, the concomitant action of epimerase and sulfotransferase could account for a higher processivity, as well as the irreversible formation of iduronic acid [130, 131, 134].
Three-dimensional structures
The crystal structure of human dermatan sulfate epimerase 1 was deposited in PDB in 2020 (PDB ID 6HZN). It contains an (α/α)6 toroid N-terminal domain, a β-sheet domain, and a C-terminal domain with a long α-helix. A manganese ion is bound close to the putative active site, which is located in a groove between the two main domains.
References
-
Tanner, Martin E. (2002) Understanding Nature's Strategies for Enzyme-Catalyzed Racemization and Epimerization. Accounts of Chemical Research vol. 35, no. 4, pp. 237-246. DOI:10.1021/ar000056y
- Van Overtveldt S, Verhaeghe T, Joosten HJ, van den Bergh T, Beerens K, and Desmet T. (2015). A structural classification of carbohydrate epimerases: From mechanistic insights to practical applications. Biotechnol Adv. 2015;33(8):1814-28. DOI:10.1016/j.biotechadv.2015.10.010 |
- Haug A and Larsen B. (1969). Biosynthesis of alginate. Epimerisation of D-mannuronic to L-guluronic acid residues in the polymer chain. Biochim Biophys Acta. 1969;192(3):557-9. DOI:10.1016/0304-4165(69)90414-0 |
-
Lindahl, Ulf and Bäckström, Gudrun and Malmström, Anders and Fransson, Lars-Åke. (1972) Biosynthesis of l-iduronic acid in heparin: Epimerization of d-glucuronic acid on the polymer level. Biochemical and Biophysical Research Communications vol. 46, no. 2, pp. 985-991. DOI:10.1016/S0006-291X(72)80238-9
- Malmström A and Fransson LA. (1975). Biosynthesis of dermatan sulfate. I. Formation of L-iduronic acid residues. J Biol Chem. 1975;250(9):3419-25. | Google Books | Open Library
-
Valla, Svein and Li, Jin-ping and Ertesvåg, Helga and Barbeyron, Tristan and Lindahl, Ulf. (2001) Hexuronyl C5-epimerases in alginate and glycosaminoglycan biosynthesis. Biochimie vol. 83, no. 8, pp. 819-830. DOI:10.1016/S0300-9084(01)01313-X
- Debarnot C, Monneau YR, Roig-Zamboni V, Delauzun V, Le Narvor C, Richard E, Hénault J, Goulet A, Fadel F, Vivès RR, Priem B, Bonnaffé D, Lortat-Jacob H, and Bourne Y. (2019). Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase. Proc Natl Acad Sci U S A. 2019;116(14):6760-6765. DOI:10.1073/pnas.1818333116 |
- Pacheco B, Maccarana M, Goodlett DR, Malmström A, and Malmström L. (2009). Identification of the active site of DS-epimerase 1 and requirement of N-glycosylation for enzyme function. J Biol Chem. 2009;284(3):1741-7. DOI:10.1074/jbc.M805479200 |
- Jerga A, Stanley MD, and Tipton PA. (2006). Chemical mechanism and specificity of the C5-mannuronan epimerase reaction. Biochemistry. 2006;45(30):9138-44. DOI:10.1021/bi060748f |
-
Gacesa, Peter. (1987) Alginate-modifying enzymes: A proposed unified mechanism of action for the lyases and epimerases. FEBS Letters vol. 212, no. 2, pp. 199-202. DOI:10.1016/0014-5793(87)81344-3
- Qin Y, Ke J, Gu X, Fang J, Wang W, Cong Q, Li J, Tan J, Brunzelle JS, Zhang C, Jiang Y, Melcher K, Li JP, Xu HE, and Ding K. (2015). Structural and functional study of D-glucuronyl C5-epimerase. J Biol Chem. 2015;290(8):4620-4630. DOI:10.1074/jbc.M114.602201 |
- Larsen B and Haug A. (1971). Biosynthesis of alginate. 1. Composition and structure of alginate produced by Azotobacter vinelandii (Lipman). Carbohydr Res. 1971;17(2):287-96. DOI:10.1016/s0008-6215(00)82536-7 |
- Haug A and Larsen B. (1971). Biosynthesis of alginate. II. Polymannuronic acid C-5-epimerase from Azotobacter vinelandii (Lipman). Carbohydr Res. 1971;17(2):297-308. DOI:10.1016/s0008-6215(00)82537-9 |
-
Stanford, Edw C C. (1883) On algin: a new substance obtained from some of the commoner species of marine algae. R. Anderson. NLM ID:101217546
-
Gorin, P. A. J. and Spencer, J. F. T. (1966) Exocellular alginic acid from Azotobacter vinelandii. Canadian Journal of Chemistry vol. 44, no. 9., pp. 993-998. DOI:10.1139/v66-147
- Linker A and Jones RS. (1966). A new polysaccharide resembling alginic acid isolated from pseudomonads. J Biol Chem. 1966;241(16):3845-51. | Google Books | Open Library
- Govan JR, Fyfe JA, and Jarman TR. (1981). Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J Gen Microbiol. 1981;125(1):217-20. DOI:10.1099/00221287-125-1-217 |
-
Okazaki, M., K. and Furuya, K. Tsukayam and K. Nisizawa. (1982) Isolation and Identification of Alginic Acid from a Calcareous Red Alga Serraticardia maxima. Botanica Marina, vol. 25, no. 3., pp. 123-131. DOI:10.1515/botm.1982.25.3.123
-
Painter, Terence J. (1983) Chapter 4 - Algal Polysaccharides. Edited by Gerald O. Aspinall. The Polysaccharides. New York: Academic Press. DOI:10.1016/B978-0-12-065602-8.50009-1
- Campos M, Martínez-Salazar JM, Lloret L, Moreno S, Núñez C, Espín G, and Soberón-Chávez G. (1996). Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178(7):1793-9. DOI:10.1128/jb.178.7.1793-1799.1996 |
- Pier GB, Coleman F, Grout M, Franklin M, and Ohman DE. (2001). Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69(3):1895-901. DOI:10.1128/IAI.69.3.1895-1901.2001 |
- Harmsen M, Yang L, Pamp SJ, and Tolker-Nielsen T. (2010). An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol. 2010;59(3):253-68. DOI:10.1111/j.1574-695X.2010.00690.x |
-
Hirst, E. L. and Jones, J. K. N and Jones, Winifred Osman. (1939) 389. The structure of alginic acid. Part I [in en]. Journal of the Chemical Society, The Royal Society of Chemistry. Vol. 0, pp. 1880–1885. DOI:10.1039/JR9390001880
- FISCHER FG and DORFEL H. (1955). [Polyuronic acids in brown algae]. Hoppe Seylers Z Physiol Chem. 1955;302(4-6):186-203. | Google Books | Open Library
-
Drummond, D W and Hirst, E L and Percival, Elizabeth. (1962) 232. The constitution of alginic acid. Journal of the Chemical Society, The Royal Society of Chemistry. Vol. 0, pp. 1208–1216. DOI:10.1039/JR9620001208
- Ertesvåg H, Høidal HK, Schjerven H, Svanem BI, and Valla S. (1999). Mannuronan C-5-epimerases and their application for in vitro and in vivo design of new alginates useful in biotechnology. Metab Eng. 1999;1(3):262-9. DOI:10.1006/mben.1999.0130 |
- Lin TY and Hassid WZ. (1966). Pathway of algnic acid synthesis in the marine brown alga, Fucus gardneri Silva. J Biol Chem. 1966;241(22):5284-97. | Google Books | Open Library
- Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, and Ohman DE. (1994). Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994;176(7):1821-30. DOI:10.1128/jb.176.7.1821-1830.1994 |
-
Haug, Arne and Larsen, Bjørn and Smidsrød, Olav. (1966) A study on the constitution of alginic acidby partial acid hydrolysis. Acta Chemica Scandinavica, vol. 5 (July), pp. 271–277. DOI:10.1016/B978-0-08-011841-3.50043-4
-
Haug, Arne and Larsen, Bjørn and Smidsrød, Olav. (1967) Studies on the Sequence of Uronic Acid Residues in Alginic Acid. Acta Chemica Scandinavica, vol. 21, pp. 691–794. DOI:10.3891/acta.chem.scand.21-0691
-
Skjåk-Bræk, Gudmund and Larsen, Bjørn and Grasdalen, Hans. (1985) The role of O-acetyl groupsin the biosynthesis of alginate by Azotobacter vinelandii. Carbohydrate Research, vol. 145, no. 1, pp. 169–174. DOI:10.1016/S0008-6215(00)90427-0
-
Kristiansen, Kåre A and Schirmer, Bjørn C and Aachmann, Finn L. and Skjåk-Bræk, Gudmund and Draget, Kurt I. and Christensen, Bjørn E. (2009) Novel alginates prepared by independent control of chain stiff-ness and distribution of G-residues: Structure and gelling properties. Carbohydrate Polymers, vol. 77, no.4, pp. 725–735. DOI:10.1016/j.carbpol.2009.02.018
- Madgwick J, Haug A, and Larsen B. (1973). Polymannuronic acid 5-epimerase from the marine alga Pelvetia canaliculata (L.) Dcne. et Thur. Acta Chem Scand. 1973;27(9):3592-4. DOI:10.3891/acta.chem.scand.27-3592 |
- Fischl R, Bertelsen K, Gaillard F, Coelho S, Michel G, Klinger M, Boyen C, Czjzek M, and Hervé C. (2016). The cell-wall active mannuronan C5-epimerases in the model brown alga Ectocarpus: From gene context to recombinant protein. Glycobiology. 2016;26(9):973-983. DOI:10.1093/glycob/cww040 |
- Nyvall P, Corre E, Boisset C, Barbeyron T, Rousvoal S, Scornet D, Kloareg B, and Boyen C. (2003). Characterization of mannuronan C-5-epimerase genes from the brown alga Laminaria digitata. Plant Physiol. 2003;133(2):726-35. DOI:10.1104/pp.103.025981 |
- Chitnis CE and Ohman DE. (1990). Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J Bacteriol. 1990;172(6):2894-900. DOI:10.1128/jb.172.6.2894-2900.1990 |
- Morea A, Mathee K, Franklin MJ, Giacomini A, O'Regan M, and Ohman DE. (2001). Characterization of algG encoding C5-epimerase in the alginate biosynthetic gene cluster of Pseudomonas fluorescens. Gene. 2001;278(1-2):107-14. DOI:10.1016/s0378-1119(01)00685-0 |
- Ertesvåg H, Doseth B, Larsen B, Skjåk-Braek G, and Valla S. (1994). Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J Bacteriol. 1994;176(10):2846-53. DOI:10.1128/jb.176.10.2846-2853.1994 |
- Ertesvåg H, Høidal HK, Hals IK, Rian A, Doseth B, and Valla S. (1995). A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol Microbiol. 1995;16(4):719-31. DOI:10.1111/j.1365-2958.1995.tb02433.x |
- Rehm BH, Ertesvåg H, and Valla S. (1996). A new Azotobacter vinelandii mannuronan C-5-epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J Bacteriol. 1996;178(20):5884-9. DOI:10.1128/jb.178.20.5884-5889.1996 |
- Svanem BI, Skjåk-Braek G, Ertesvåg H, and Valla S. (1999). Cloning and expression of three new Aazotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5-epimerases. J Bacteriol. 1999;181(1):68-77. DOI:10.1128/JB.181.1.68-77.1999 |
- Gawin A, Tietze L, Aarstad OA, Aachmann FL, Brautaset T, and Ertesvåg H. (2020). Functional characterization of three Azotobacter chroococcum alginate-modifying enzymes related to the Azotobacter vinelandii AlgE mannuronan C-5-epimerase family. Sci Rep. 2020;10(1):12470. DOI:10.1038/s41598-020-68789-3 |
- Gimmestad M, Sletta H, Ertesvåg H, Bakkevig K, Jain S, Suh SJ, Skjåk-Braek G, Ellingsen TE, Ohman DE, and Valla S. (2003). The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J Bacteriol. 2003;185(12):3515-23. DOI:10.1128/JB.185.12.3515-3523.2003 |
- Ertesvåg H and Valla S. (1999). The A modules of the Azotobacter vinelandii mannuronan-C-5-epimerase AlgE1 are sufficient for both epimerization and binding of Ca2+. J Bacteriol. 1999;181(10):3033-8. DOI:10.1128/JB.181.10.3033-3038.1999 |
- Ertesvåg H (2015). Alginate-modifying enzymes: biological roles and biotechnological uses. Front Microbiol. 2015;6:523. DOI:10.3389/fmicb.2015.00523 |
-
Ramstad, Marit Valeur and Ellingsen, Trond E. and Josefsen, Kjell D. and Høidal, Hilde K. and Valla, Svein and Skjåk-Bræk, Gudmund and Levine, David W. (1999) Properties and action pattern of the recombinant mannuronan C-5-epimerase AlgE2. Enzyme and Microbial Technology, vol. 24, no. 10, pp. 636–646. DOI:10.1016/S0141-0229(98)00148-3
- Holtan S, Bruheim P, and Skjåk-Braek G. (2006). Mode of action and subsite studies of the guluronan block-forming mannuronan C-5 epimerases AlgE1 and AlgE6. Biochem J. 2006;395(2):319-29. DOI:10.1042/BJ20051804 |
- Buchinger E, Knudsen DH, Behrens MA, Pedersen JS, Aarstad OA, Tøndervik A, Valla S, Skjåk-Bræk G, Wimmer R, and Aachmann FL. (2014). Structural and functional characterization of the R-modules in alginate C-5 epimerases AlgE4 and AlgE6 from Azotobacter vinelandii. J Biol Chem. 2014;289(45):31382-96. DOI:10.1074/jbc.M114.567008 |
- Buchinger E, Knudsen DH, Behrens MA, Pedersen JS, Aarstad OA, Tøndervik A, Valla S, Skjåk-Bræk G, Wimmer R, and Aachmann FL. (2014). Structural and functional characterization of the R-modules in alginate C-5 epimerases AlgE4 and AlgE6 from Azotobacter vinelandii. J Biol Chem. 2014;289(45):31382-96. DOI:10.1074/jbc.M114.567008 |
- Campa C, Holtan S, Nilsen N, Bjerkan TM, Stokke BT, and Skjåk-Braek G. (2004). Biochemical analysis of the processive mechanism for epimerization of alginate by mannuronan C-5 epimerase AlgE4. Biochem J. 2004;381(Pt 1):155-64. DOI:10.1042/BJ20031265 |
- Høidal HK, Ertesvåg H, Skjåk-Braek G, Stokke BT, and Valla S. (1999). The recombinant Azotobacter vinelandii mannuronan C-5-epimerase AlgE4 epimerizes alginate by a nonrandom attack mechanism. J Biol Chem. 1999;274(18):12316-22. DOI:10.1074/jbc.274.18.12316 |
- Hartmann M, Holm OB, Johansen GA, Skjåk-Braek G, and Stokke BT. (2002). Mode of action of recombinant Azotobacter vinelandii mannuronan C-5 epimerases AlgE2 and AlgE4. Biopolymers. 2002;63(2):77-88. DOI:10.1002/bip.10017 |
-
Sletmoen, Marit and Skjåk-Bræk, Gudmund and Stokke, Bjørn T. (2004) Single-molecular Pair Unbinding Studies of Mannuronan C-5 Epimerase AlgE4 and Its Polymer Substrate, Biomacromolecules, American Chemical Society, vol. 5, no.4, pp. 1288–1295. DOI:10.1021/BM0345211
- Håti AG, Aachmann FL, Stokke BT, Skjåk-Bræk G, and Sletmoen M. (2015). Energy Landscape of Alginate-Epimerase Interactions Assessed by Optical Tweezers and Atomic Force Microscopy. PLoS One. 2015;10(10):e0141237. DOI:10.1371/journal.pone.0141237 |
- Svanem BI, Strand WI, Ertesvag H, Skjåk-Braek G, Hartmann M, Barbeyron T, and Valla S. (2001). The catalytic activities of the bifunctional Azotobacter vinelandii mannuronan C-5-epimerase and alginate lyase AlgE7 probably originate from the same active site in the enzyme. J Biol Chem. 2001;276(34):31542-50. DOI:10.1074/jbc.M102562200 |
-
Yoon, Hye-Jin and Hashimoto, Wataru and Miyake, Osamu and Murata, Kousaku and Mikami, Bunzo. (2001) Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution, Journal of Molecular Biology, vol. 307, no. 1 pp. 9–16. DOI:10.1006/jmbi.2000.4509
- Ertesvåg H, Erlien F, Skjåk-Braek G, Rehm BH, and Valla S. (1998). Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J Bacteriol. 1998;180(15):3779-84. DOI:10.1128/JB.180.15.3779-3784.1998 |
- DISCHE Z (1947). A new specific color reaction of hexuronic acids. J Biol Chem. 1947;167(1):189-98. | Google Books | Open Library
- Knutson CA and Jeanes A. (1968). A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968;24(3):470-81. DOI:10.1016/0003-2697(68)90154-1 |
- Knutson CA and Jeanes A. (1968). Determination of the composition of uronic acid mixtures. Anal Biochem. 1968;24(3):482-90. DOI:10.1016/0003-2697(68)90155-3 |
-
Skjåk-Bræk, Gudmund, and Larsen, Bjørn. (1982) Biosynthesis of Alginate Part 5. A New Assay for Mannuronan C-5-Epimerase Activity, Carbohydrate Research, vol. 103, no.1, pp. 133–136. DOI:10.1016/S0008-6215(82)80013-X
-
Skjåk-Bræk, Gudmund and Larsen, Bjørn. (1985) Biosynthesis of alginate: Purification and characterisation of mannuronan C-5-epimerase from Azotobacter vinelandii, Carbohydrate Research, vol. 139 (June), pp. 273–283. DOI:10.1016/0008-6215(85)90026-6
-
Currie, Andrew J., and Turvey, James R. (1982) An enzymic method for the assay of d-mannuronanC-5 epimerase activity. Carbohydrate Research, vol. 107, no. 1, pp. 156–159. DOI:10.1016/S0008-6215(00)80786-7
- PREISS J and ASHWELL G. (1962). Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. J Biol Chem. 1962;237:309-16. | Google Books | Open Library
- Boyd J and Turvey JR. (1977). Isolation of poly-alpha-L-guluronate lyase from Klebsiella aerogenes. Carbohydr Res. 1977;57:163-71. DOI:10.1016/s0008-6215(00)81928-x |
-
Grasdalen, Hans and Larsen, Bjørn and Smidsrød, Olav. (1977) 13C-N.m.r. studies of alginate. Carbohydrate Research, vol. 56, no. 2, pp. C11–C15. DOI:10.1016/S0008-6215(00)83369-8
-
Grasdalen, Hans and Larsen, Bjørn and Smidsrød, Olav. (1981) 13C-n.m.r. studies of monomeric composition and sequence in alginate. Carbohydrate Research, vol. 89, no. 2, pp. 179–191. DOI:10.1016/S0008-6215(00)85243-X
-
Grasdalen, Hans and Larsen, Bjørn and Smidsrød, Olav. (1979) A p.m.r. study of the composition and sequence of uronate residues in alginates. Carbohydrate Research, vol. 68, no. 1, pp. 23–31. DOI:10.1016/S0008-6215(00)84051-3
-
Grasdalen, Hans. (1983) High-field, 1H-n.m.r. spectroscopy of alginate: sequential structure andlinkage conformations. Carbohydrate Research, vol. 118 (July), pp. 255–260. DOI:10.1016/0008-6215(83)88053-7
- Hartmann M, Duun AS, Markussen S, Grasdalen H, Valla S, and Skjåk-Braek G. (2002). Time-resolved 1H and 13C NMR spectroscopy for detailed analyses of the Azotobacter vinelandii mannuronan C-5 epimerase reaction. Biochim Biophys Acta. 2002;1570(2):104-12. DOI:10.1016/s0304-4165(02)00195-2 |
- Tøndervik A, Klinkenberg G, Aachmann FL, Svanem BI, Ertesvåg H, Ellingsen TE, Valla S, Skjåk-Bræk G, and Sletta H. (2013). Mannuronan C-5 epimerases suited for tailoring of specific alginate structures obtained by high-throughput screening of an epimerase mutant library. Biomacromolecules. 2013;14(8):2657-66. DOI:10.1021/bm4005194 |
- Stanisci A, Tøndervik A, Gaardløs M, Lervik A, Skjåk-Bræk G, Sletta H, and Aachmann FL. (2020). Identification of a Pivotal Residue for Determining the Block Structure-Forming Properties of Alginate C-5 Epimerases. ACS Omega. 2020;5(8):4352-4361. DOI:10.1021/acsomega.9b04490 |
-
Morris, Edwin R., Rees, David A. and Thom, David. (1973) Characterization of polysaccharide structure and interactions by circular dichroism: order–disorder transition in the calcium alginate system. Journal of the Chemical Society, The Royal Society of Chemistry, Chemical Communications, no. 7, pp. 245–246. DOI:10.1039/C39730000245
-
Donati, Ivan and Gamini, Amelia and Skjåk-Bræk, Gudmund and Vetere, Amedeo and Campa, Cristiana and Coslovi, Anna and Paoletti, Sergio. (2003) Determination of the diadic composition of alginate by means of circular dichroism: a fast and accurate improved method. Carbohydrate Research, vol. 338, no. 10, pp. 1139–1142. DOI:10.1016/S0008-6215(03)00094-6
-
Reese, Johanna de and Sperl, Nadine and Schmid, Jochen and Sieber, Volker and Plank, Johann. (2015) Effect of biotechnologically modified alginates on LDH structures. Bioinspired, Biomimetic and Nanobiomaterials, vol. 4, no. 3, pp. 174–186. DOI:10.1680/jbibn.14.00032
- Tøndervik A, Klinkenberg G, Aarstad OA, Drabløs F, Ertesvåg H, Ellingsen TE, Skjåk-Bræk G, Valla S, and Sletta H. (2010). Isolation of mutant alginate lyases with cleavage specificity for di-guluronic acid linkages. J Biol Chem. 2010;285(46):35284-92. DOI:10.1074/jbc.M110.162800 |
- Campa C, Oust A, Skjåk-Braek G, Paulsen BS, Paoletti S, Christensen BE, and Ballance S. (2004). Determination of average degree of polymerisation and distribution of oligosaccharides in a partially acid-hydrolysed homopolysaccharide: a comparison of four experimental methods applied to mannuronan. J Chromatogr A. 2004;1026(1-2):271-81. DOI:10.1016/j.chroma.2003.11.045 |
-
Ballance, Simon and Holtan, Synnøve and Aarstad, Olav Andreas and Sikorski, Pawel and Skjåk-Bræk, Gudmund and Christensen, Bjørn E. (2005) Application of high-performance anion-exchange chromatography with pulsed amperometric detection and statistical analysis to study oligosaccharide distributions – a complementary method to investigate the structure and some properties of alginates. Journal of Chromatography A, vol. 1093, no. 1, pp. 59–68. DOI:10.1016/j.chroma.2005.07.051
- Aarstad OA, Tøndervik A, Sletta H, and Skjåk-Bræk G. (2012). Alginate sequencing: an analysis of block distribution in alginates using specific alginate degrading enzymes. Biomacromolecules. 2012;13(1):106-16. DOI:10.1021/bm2013026 |
- Rozeboom HJ, Bjerkan TM, Kalk KH, Ertesvåg H, Holtan S, Aachmann FL, Valla S, and Dijkstra BW. (2008). Structural and mutational characterization of the catalytic A-module of the mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii. J Biol Chem. 2008;283(35):23819-28. DOI:10.1074/jbc.M804119200 |
- Wolfram F, Kitova EN, Robinson H, Walvoort MT, Codée JD, Klassen JS, and Howell PL. (2014). Catalytic mechanism and mode of action of the periplasmic alginate epimerase AlgG. J Biol Chem. 2014;289(9):6006-19. DOI:10.1074/jbc.M113.533158 |
-
Jurrus, Elizabeth and Engel, Dave and Star, Keith and Monson, Kyle and Brandi, Juan and Felberg, Lisa E. and Brookes, David H. et al. (2018) Improvements to the APBS biomolecular solvation software suite. Protein Science, vol. 27, no. 1, pp. 112–128. DOI:10.1002/pro.3280
-
Schrödinger, LLC. (2018) The PyMOL Molecular Graphics System, Version 2.2.3.
- Douthit SA, Dlakic M, Ohman DE, and Franklin MJ. (2005). Epimerase active domain of Pseudomonas aeruginosa AlgG, a protein that contains a right-handed beta-helix. J Bacteriol. 2005;187(13):4573-83. DOI:10.1128/JB.187.13.4573-4583.2005 |
-
Ofstad, Ragni and Larsen, Bjørn. (1981) The effect of calcium ion concentration on poly-D-mannuronate C-5 epimerase. Proceedings of the 10th International Seaweed Symposium, edited by Tore Levring, pp. 485–493.
- Ertesvåg H, Høidal HK, Skjåk-Braek G, and Valla S. (1998). The Azotobacter vinelandii mannuronan C-5-epimerase AlgE1 consists of two separate catalytic domains. J Biol Chem. 1998;273(47):30927-32. DOI:10.1074/jbc.273.47.30927 |
- Huang W, Matte A, Li Y, Kim YS, Linhardt RJ, Su H, and Cygler M. (1999). Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution. J Mol Biol. 1999;294(5):1257-69. DOI:10.1006/jmbi.1999.3292 |
- Michel G, Pojasek K, Li Y, Sulea T, Linhardt RJ, Raman R, Prabhakar V, Sasisekharan R, and Cygler M. (2004). The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J Biol Chem. 2004;279(31):32882-96. DOI:10.1074/jbc.M403421200 |
- Xu F, Dong F, Wang P, Cao HY, Li CY, Li PY, Pang XH, Zhang YZ, and Chen XL. (2017). Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6. J Biol Chem. 2017;292(11):4457-4468. DOI:10.1074/jbc.M116.766030 |
- Stender EGP, Dybdahl Andersen C, Fredslund F, Holck J, Solberg A, Teze D, Peters GHJ, Christensen BE, Aachmann FL, Welner DH, and Svensson B. (2019). Structural and functional aspects of mannuronic acid-specific PL6 alginate lyase from the human gut microbe Bacteroides cellulosilyticus. J Biol Chem. 2019;294(47):17915-17930. DOI:10.1074/jbc.RA119.010206 |
- Aachmann FL, Svanem BI, Güntert P, Petersen SB, Valla S, and Wimmer R. (2006). NMR structure of the R-module: a parallel beta-roll subunit from an Azotobacter vinelandii mannuronan C-5 epimerase. J Biol Chem. 2006;281(11):7350-6. DOI:10.1074/jbc.M510069200 |
-
Sletmoen, Marit and Skjåk-Bræk, Gudmund and Stokke, Bjørn T. (2005) Mapping enzymatic functionalities of mannuronan C-5 epimerases and their modular units by dynamic force spectroscopy. Carbohydrate Research, vol. 340, no.18, pp. 2782–2795. DOI:10.1016/J.CARRES.2005.09.020
- Yoder MD, Lietzke SE, and Jurnak F. (1993). Unusual structural features in the parallel beta-helix in pectate lyases. Structure. 1993;1(4):241-51. DOI:10.1016/0969-2126(93)90013-7 |
- Welch RA (1991). Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5(3):521-8. DOI:10.1111/j.1365-2958.1991.tb00723.x |
-
Lilie, Hauke and Haehnel, Wolfgang and Rudolph, Rainer and Baumann, Ulrich. (2000) Folding of a synthetic parallel β-roll protein. FEBS Letters, vol. 470, no. 2, pp. 173–177. DOI:10.1016/S0014-5793(00)01308-9
-
Jenkins, John and Pickersgill, Richard. (2001) The architecture of parallel β-helices and related folds. Progress in Biophysics and Molecular Biology, vol. 77, no. 2, pp. 111–175. DOI:10.1016/S0079-6107(01)00013-X
- Gimmestad M, Steigedal M, Ertesvåg H, Moreno S, Christensen BE, Espín G, and Valla S. (2006). Identification and characterization of an Azotobacter vinelandii type I secretion system responsible for export of the AlgE-type mannuronan C-5-epimerases. J Bacteriol. 2006;188(15):5551-60. DOI:10.1128/JB.00236-06 |
- Yoder MD, Keen NT, and Jurnak F. (1993). New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260(5113):1503-7. DOI:10.1126/science.8502994 |
- Kajava AV and Steven AC. (2006). Beta-rolls, beta-helices, and other beta-solenoid proteins. Adv Protein Chem. 2006;73:55-96. DOI:10.1016/S0065-3233(06)73003-0 |
- Rigden DJ and Franco OL. (2002). Beta-helical catalytic domains in glycoside hydrolase families 49, 55 and 87: domain architecture, modelling and assignment of catalytic residues. FEBS Lett. 2002;530(1-3):225-32. DOI:10.1016/s0014-5793(02)03490-7 |
-
Garron, M.-L. and Cygler, M. (2010) Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology, vol. 20, no. 12, pp. 1547–1573. DOI:10.1093/glycob/cwq122
-
Larsson, Anna M and Andersson, Rolf and Ståhlberg, Jerry and Kenne, Lennart and Jones, T. Alwyn. (2003) Dextranase from Penicillium minioluteum: Reaction Course, Crystal Structure, and Product Complex. Structure, vol. 11, no. 9, pp. 1111–1121. DOI:10.1016/S0969-2126(03)00147-3
- Petersen TN, Kauppinen S, and Larsen S. (1997). The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel beta helix. Structure. 1997;5(4):533-44. DOI:10.1016/s0969-2126(97)00209-8 |
- Pickersgill R, Smith D, Worboys K, and Jenkins J. (1998). Crystal structure of polygalacturonase from Erwinia carotovora ssp. carotovora. J Biol Chem. 1998;273(38):24660-4. DOI:10.1074/jbc.273.38.24660 |
- Ishida T, Fushinobu S, Kawai R, Kitaoka M, Igarashi K, and Samejima M. (2009). Crystal structure of glycoside hydrolase family 55 {beta}-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. J Biol Chem. 2009;284(15):10100-9. DOI:10.1074/jbc.M808122200 |
- Michel G, Chantalat L, Fanchon E, Henrissat B, Kloareg B, and Dideberg O. (2001). The iota-carrageenase of Alteromonas fortis. A beta-helix fold-containing enzyme for the degradation of a highly polyanionic polysaccharide. J Biol Chem. 2001;276(43):40202-9. DOI:10.1074/jbc.M100670200 |
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, and Reinemer P. (1994). Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science. 1994;265(5170):383-6. DOI:10.1126/science.8023158 |
-
Mayans, O. and Scott, M. and Connerton, I. and Gravesen, T. and Benen, J. and Visser, J. and Pickersgill, R. and Jenkins, J. (1997) Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure, vol. 5, no. 5, pp. 677–689. DOI:10.1016/S0969-2126(97)00222-0
- Akita M, Suzuki A, Kobayashi T, Ito S, and Yamane T. (2001). The first structure of pectate lyase belonging to polysaccharide lyase family 3. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 12):1786-92. DOI:10.1107/s0907444901014482 |
- Jenkins J, Shevchik VE, Hugouvieux-Cotte-Pattat N, and Pickersgill RW. (2004). The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi. J Biol Chem. 2004;279(10):9139-45. DOI:10.1074/jbc.M311390200 |
-
Jenkins, John and Mayans, Olga and Pickersgill, Richard. (1998) Structure and Evolution of Parallel β-Helix Proteins. Journal of Structural Biology, vol. 122, nos. 1-2, pp. 236–246. DOI:10.1006/JSBI.1998.3985
- Höök M, Lindahl U, Bäckström G, Malmström A, and Fransson L. (1974). Biosynthesis of heparin. 3. Formation of iduronic acid residues. J Biol Chem. 1974;249(12):3908-15. | Google Books | Open Library
- Gallagher JT and Walker A. (1985). Molecular distinctions between heparan sulphate and heparin. Analysis of sulphation patterns indicates that heparan sulphate and heparin are separate families of N-sulphated polysaccharides. Biochem J. 1985;230(3):665-74. DOI:10.1042/bj2300665 |
- Bishop JR, Schuksz M, and Esko JD. (2007). Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446(7139):1030-7. DOI:10.1038/nature05817 |
- Jacobsson I, Lindahl U, Jensen JW, Rodén L, Prihar H, and Feingold DS. (1984). Biosynthesis of heparin. Substrate specificity of heparosan N-sulfate D-glucuronosyl 5-epimerase. J Biol Chem. 1984;259(2):1056-63. | Google Books | Open Library
-
Hagner-McWhirter, Åsa and Hannesson, Helgi H. and Campbell, Patrick and Westley, John and Rodén, Lennart and Lindahl, Ulf and Li, Jin-Ping. (2000) Biosynthesis of heparin/heparan sulfate: kinetic studies of the glucuronyl C5-epimerase with N-sulfated derivatives of the Escherichia coli K5 capsular polysaccharide as substrates. Glycobiology vol. 10, no. 2, pp. 159-171. DOI:10.1093/glycob/10.2.159
- Hagner-McWhirter A, Li JP, Oscarson S, and Lindahl U. (2004). Irreversible glucuronyl C5-epimerization in the biosynthesis of heparan sulfate. J Biol Chem. 2004;279(15):14631-8. DOI:10.1074/jbc.M313760200 |
- Li JP (2010). Glucuronyl C5-epimerase an enzyme converting glucuronic acid to iduronic acid in heparan sulfate/heparin biosynthesis. Prog Mol Biol Transl Sci. 2010;93:59-78. DOI:10.1016/S1877-1173(10)93004-4 |
- Sheng J, Xu Y, Dulaney SB, Huang X, and Liu J. (2012). Uncovering biphasic catalytic mode of C5-epimerase in heparan sulfate biosynthesis. J Biol Chem. 2012;287(25):20996-1002. DOI:10.1074/jbc.M112.359885 |
- Mochizuki H, Yamagishi K, Suzuki K, Kim YS, and Kimata K. (2015). Heparosan-glucuronate 5-epimerase: Molecular cloning and characterization of a novel enzyme. Glycobiology. 2015;25(7):735-44. DOI:10.1093/glycob/cwv013 |
- Han YH, Garron ML, Kim HY, Kim WS, Zhang Z, Ryu KS, Shaya D, Xiao Z, Cheong C, Kim YS, Linhardt RJ, Jeon YH, and Cygler M. (2009). Structural snapshots of heparin depolymerization by heparin lyase I. J Biol Chem. 2009;284(49):34019-27. DOI:10.1074/jbc.M109.025338 |
- Lunin VV, Li Y, Linhardt RJ, Miyazono H, Kyogashima M, Kaneko T, Bell AW, and Cygler M. (2004). High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism. J Mol Biol. 2004;337(2):367-86. DOI:10.1016/j.jmb.2003.12.071 |
- Prihar HS, Campbell P, Feingold DS, Jacobsson I, Jensen JW, Lindahl U, and Rodén L. (1980). Biosynthesis of heparin. Hydrogen exchange at carbon 5 of the glucuronosyl residues. Biochemistry. 1980;19(3):495-500. DOI:10.1021/bi00544a016 |
- Hagner-Mcwhirter A, Lindahl U, and Li Jp. (2000). Biosynthesis of heparin/heparan sulphate: mechanism of epimerization of glucuronyl C-5. Biochem J. 2000;347 Pt 1(Pt 1):69-75. | Google Books | Open Library
- Malmström A, Rodén L, Feingold DS, Jacobsson I, Bäckström G, and Lindahl U. (1980). Biosynthesis of heparin. Partial purification of the uronosyl C-5 epimerase. J Biol Chem. 1980;255(9):3878-83. | Google Books | Open Library
- Malmström A and Aberg L. (1982). Biosynthesis of dermatan sulphate. Assay and properties of the uronosyl C-5 epimerase. Biochem J. 1982;201(3):489-93. DOI:10.1042/bj2010489 |
- Maccarana M, Olander B, Malmström J, Tiedemann K, Aebersold R, Lindahl U, Li JP, and Malmström A. (2006). Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. J Biol Chem. 2006;281(17):11560-8. DOI:10.1074/jbc.M513373200 |
- Pacheco B, Malmström A, and Maccarana M. (2009). Two dermatan sulfate epimerases form iduronic acid domains in dermatan sulfate. J Biol Chem. 2009;284(15):9788-95. DOI:10.1074/jbc.M809339200 |
- Trowbridge JM and Gallo RL. (2002). Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology. 2002;12(9):117R-25R. DOI:10.1093/glycob/cwf066 |
-
US National Library of Medicine. (2020) Chondroitin Sulfates MeSH Descriptor Data 2020. Unique ID: D002809
- Malmström A (1984). Biosynthesis of dermatan sulfate. II. Substrate specificity of the C-5 uronosyl epimerase. J Biol Chem. 1984;259(1):161-5. | Google Books | Open Library
- Tykesson E, Hassinen A, Zielinska K, Thelin MA, Frati G, Ellervik U, Westergren-Thorsson G, Malmström A, Kellokumpu S, and Maccarana M. (2018). Dermatan sulfate epimerase 1 and dermatan 4-O-sulfotransferase 1 form complexes that generate long epimerized 4-O-sulfated blocks. J Biol Chem. 2018;293(35):13725-13735. DOI:10.1074/jbc.RA118.003875 |
- Tykesson E, Mao Y, Maccarana M, Pu Y, Gao J, Lin C, Zaia J, Westergren-Thorsson G, Ellervik U, Malmström L, and Malmström A. (2016). Deciphering the Mode of Action of the Processive Polysaccharide Modifying Enzyme Dermatan Sulfate Epimerase 1 by Hydrogen-Deuterium Exchange Mass Spectrometry. Chem Sci. 2016;7(2):1447-1456. DOI:10.1039/C5SC03798K |
- Hannesson HH, Hagner-McWhirter A, Tiedemann K, Lindahl U, and Malmström A. (1996). Biosynthesis of dermatan sulphate. Defructosylated Escherichia coli K4 capsular polysaccharide as a substrate for the D-glucuronyl C-5 epimerase, and an indication of a two-base reaction mechanism. Biochem J. 1996;313 ( Pt 2)(Pt 2):589-96. DOI:10.1042/bj3130589 |
- Eklund E, Rodén L, Malmström M, and Malmström A. (2000). Dermatan is a better substrate for 4-O-sulfation than chondroitin: implications in the generation of 4-O-sulfated, L-iduronate-rich galactosaminoglycans. Arch Biochem Biophys. 2000;383(2):171-7. DOI:10.1006/abbi.2000.2043 |