CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Esterase Family 3"
Joel Weadge (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | * [[Author]]: | + | * [[Author]]: [[User:Stefen Stangherlin|Stefen Stangherlin]] |
| − | * [[Responsible Curator]]s: | + | * [[Responsible Curator]]s: [[User:Michael Suits|Michael Suits]] and [[User:Joel Weadge|Joel Weadge]] |
---- | ---- | ||
| Line 11: | Line 11: | ||
|{{Hl2}} colspan="2" align="center" |'''Carbohydrate Esterase Family CE3''' | |{{Hl2}} colspan="2" align="center" |'''Carbohydrate Esterase Family CE3''' | ||
|- | |- | ||
| − | |''' | + | |'''Fold''' |
|(α/β/α)-Sandwich | |(α/β/α)-Sandwich | ||
|- | |- | ||
| Line 40: | Line 40: | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The CE3 family has a number of enzymes that have been structurally resolved | + | The CE3 family has a number of enzymes that have been structurally resolved. Examples include ''Tc''AE206 from ''Talaromyces cellulolyticus'' ([{{PDBlink}}5B5S PDB ID 5B5S]) (see Fig. 1) and ''Ct''Ces3-1 from ''Hungateiclostridium thermocellum'' (formerly ''Clostridium thermocellum'') ([{{PDBlink}}2VPT PDB ID 2VPT]). Both structures adopt an (α/β/α)-sandwich fold typical of the SGNH hydrolase family. The (α/β/α)-sandwich contains five central parallel β-strands forming a curved β-sheet, which is flanked by 5-6 α-helices <cite>Correia2008 Uechi2016</cite>. Additionally, both structures contain a calcium binding loop motif (DXVGX<sub>7</sub>DX<sub>n</sub>(D/N)) located above the N-terminal end of the central β-strand (β2) <cite>Watanabe2015</cite>. This binding motif is conserved across all currently characterized CE3s. A coordinated zinc ion was also observed next to a calcium ion in a ''Tc''AE206_S10A variant ([{{PDBlink}}5B5L PDB ID 5B5L]), however this was attributed to the use of ZnSO<sub>4</sub> in the crystallization conditions <cite>Uechi2016</cite>. Unique to ''Tc''AE206 is a disulfide bond formed near the N-terminus (see Fig. 1) that is thought to position the catalytic Ser by stabilizing neighbouring areas, including a β-turn (β1) that involves the catalytic Ser <cite>Uechi2016 Watanabe2015</cite>. |
== Family Firsts == | == Family Firsts == | ||
Latest revision as of 13:16, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Carbohydrate Esterase Family CE3 | |
| Fold | (α/β/α)-Sandwich |
| Mechanism | Serine Hydrolase |
| Active site residues | Known, Catalytic Triad |
| CAZy DB link | |
| https://www.cazy.org/CE3.html | |
Substrate specificities
Carbohydrate Esterase Family 3 is currently comprised entirely of de-O-acetylxylan esterases. Xylan is a plant cell-wall polysaccharide composed of β-1,4-linked xylose decorated with α-arabinofuranose and α-glucuronic acid substituents [1].
Catalytic Residues
Functionally characterized CE3 members are all known to contain the classical catalytic triad of Ser-His-Asp, typical of the SGNH hydrolase family of enzymes (see Fig. 1) [2, 3]. The active site residues are presented via four conserved consensus sequences (Blocks I-III and V), and have an altered nucleophilic “elbow” turn motif (-GxSxT- as opposed to the canonical -GxSxG- motif) compared to other related members of the α/β-hydrolase family [4, 5]. The catalytic triad along with the Block II Gly and Block III Asn residues that comprise the oxyanion hole, are universally conserved across all characterized CE3 enzymes. The Block V Asp residue mediates the amphoteric nature of the Block V His residue, which abstracts a proton from the Block I Ser to render it nucleophilic.
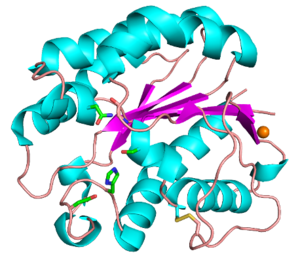
Kinetics and Mechanism
CE3 esterases catalyze the hydrolysis of O-linked acetyl groups from xylan oligo- and poly-saccharides. The Block V His residue abstracts a proton from the Block I Ser, rendering it nucleophilic, which attacks the electrophilic carbonyl carbon of the acetyl group of the xylan substrate; generating a tetrahedral oxyanion intermediate that is stabilized by the backbone amides of the Block I Ser and Block II Gly, as well as the sidechain amide of the Block III Asn, together forming the oxyanion hole in the active site [6]. Collapse of the oxyanion intermediate results in the formation of a transient acyl-enzyme intermediate and alcohol by-product [6]. A hydrolytic water molecule is then deprotonated by the Block V His residue, and attacks the acyl-enzyme intermediate; hydrolyzing the bond and releasing acetate and the free enzyme [6]. In the process, the Ser (Block I) is re-protonated and ready for another catalytic cycle.
The kinetics for enzymes from the CE3 family have been examined. For example, CtCes3-1 (PDB ID 2VPT) was found to have a kcat/KM of 2.5 and 1.2 mM-1s-1 for p-nitrophenyl acetate (pNP-Ac) and acetylated xylan, respectively [7]. In other studies, TcAE206 (PDB ID 5B5S) was not assayed against acetylated xylan, but the kcat/KM with pNP-Ac and p-nitrophenyl butyrate (pNP-B) were reported as 44.7 and 4.1 mM-1s-1, respectively, while no activity was detected with p‐nitrophenyl octanoate (pNP-O) as the substrate [8].
Three-dimensional structures
The CE3 family has a number of enzymes that have been structurally resolved. Examples include TcAE206 from Talaromyces cellulolyticus (PDB ID 5B5S) (see Fig. 1) and CtCes3-1 from Hungateiclostridium thermocellum (formerly Clostridium thermocellum) (PDB ID 2VPT). Both structures adopt an (α/β/α)-sandwich fold typical of the SGNH hydrolase family. The (α/β/α)-sandwich contains five central parallel β-strands forming a curved β-sheet, which is flanked by 5-6 α-helices [6, 7]. Additionally, both structures contain a calcium binding loop motif (DXVGX7DXn(D/N)) located above the N-terminal end of the central β-strand (β2) [8]. This binding motif is conserved across all currently characterized CE3s. A coordinated zinc ion was also observed next to a calcium ion in a TcAE206_S10A variant (PDB ID 5B5L), however this was attributed to the use of ZnSO4 in the crystallization conditions [6]. Unique to TcAE206 is a disulfide bond formed near the N-terminus (see Fig. 1) that is thought to position the catalytic Ser by stabilizing neighbouring areas, including a β-turn (β1) that involves the catalytic Ser [6, 8].
Family Firsts
- First characterized
- In 1994, the sequence of XynB from Ruminococcus flavefaciens 17 was found to be related to family G xylanases [9]. In 1997, BnaC from Neocallimastix patriciarum was found to have close relation to XynB and other enzymes known to be members of a diverse family of esterases [10]. It wasn’t until 2000 that CesA from R. flavefaciens 17, which was shown to have significant sequence identity to XynB, was characterized with the ability to deacetylate acetylated xylans; thereby representing the first characterized enzymes of family 3 CEs [11].
- First mechanistic insight
- In 2000, CesA, XynB, and BnaC were aligned and shown to contain what was thought to be a Ser-His-Asp catalytic triad responsible for the observed esterase activity [11]. This was later confirmed by the structural resolution of CtCes3-1 (PDB ID 2VPT) [7].
- First 3-D structure
- The first resolved structure was CtCes3-1 (PDB ID 2VPT) from Hungateiclostridium thermocellum (formerly Clostridium thermocellum), displaying the (α/β/α)-sandwich fold and Ser-His-Asp catalytic triad typical of SGNH hydrolases [7].
References
- Faik A (2010). Xylan biosynthesis: news from the grass. Plant Physiol. 2010;153(2):396-402. DOI:10.1104/pp.110.154237 |
- Polgár L (2005). The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005;62(19-20):2161-72. DOI:10.1007/s00018-005-5160-x |
- Mølgaard A, Kauppinen S, and Larsen S. (2000). Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure. 2000;8(4):373-83. DOI:10.1016/s0969-2126(00)00118-0 |
- Upton C and Buckley JT. (1995). A new family of lipolytic enzymes?. Trends Biochem Sci. 1995;20(5):178-9. DOI:10.1016/s0968-0004(00)89002-7 |
- Akoh CC, Lee GC, Liaw YC, Huang TH, and Shaw JF. (2004). GDSL family of serine esterases/lipases. Prog Lipid Res. 2004;43(6):534-52. DOI:10.1016/j.plipres.2004.09.002 |
- Uechi K, Kamachi S, Akita H, Mine S, and Watanabe M. (2016). Crystal structure of an acetyl esterase complexed with acetate ion provides insights into the catalytic mechanism. Biochem Biophys Res Commun. 2016;477(3):383-7. DOI:10.1016/j.bbrc.2016.06.093 |
- Correia MA, Prates JA, Brás J, Fontes CM, Newman JA, Lewis RJ, Gilbert HJ, and Flint JE. (2008). Crystal structure of a cellulosomal family 3 carbohydrate esterase from Clostridium thermocellum provides insights into the mechanism of substrate recognition. J Mol Biol. 2008;379(1):64-72. DOI:10.1016/j.jmb.2008.03.037 |
- Watanabe M, Fukada H, Inoue H, and Ishikawa K. (2015). Crystal structure of an acetylesterase from Talaromyces cellulolyticus and the importance of a disulfide bond near the active site. FEBS Lett. 2015;589(11):1200-6. DOI:10.1016/j.febslet.2015.03.020 |
- Zhang JX, Martin J, and Flint HJ. (1994). Identification of non-catalytic conserved regions in xylanases encoded by the xynB and xynD genes of the cellulolytic rumen anaerobe Ruminococcus flavefaciens. Mol Gen Genet. 1994;245(2):260-4. DOI:10.1007/BF00283275 |
- Dalrymple BP, Cybinski DH, Layton I, McSweeney CS, Xue GP, Swadling YJ, and Lowry JB. (1997). Three Neocallimastix patriciarum esterases associated with the degradation of complex polysaccharides are members of a new family of hydrolases. Microbiology (Reading). 1997;143 ( Pt 8):2605-2614. DOI:10.1099/00221287-143-8-2605 |
- Aurilia V, Martin JC, McCrae SI, Scott KP, Rincon MT, and Flint HJ. (2000). Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology (Reading). 2000;146 ( Pt 6):1391-1397. DOI:10.1099/00221287-146-6-1391 |