CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Polysaccharide Lyase Family 1"
Wade Abbott (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (48 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Richard Pickersgill|Richard Pickersgill]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Richard Pickersgill|Richard Pickersgill]] |
---- | ---- | ||
| Line 29: | Line 29: | ||
</div> | </div> | ||
<!-- This is the end of the table --> | <!-- This is the end of the table --> | ||
| + | |||
== Substrate specificities == | == Substrate specificities == | ||
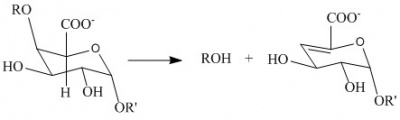
| − | [[Image:Reaction.jpg|thumb|400px|'''Figure 1.''' | + | [[Image:Reaction.jpg|thumb|400px|'''Figure 1.''' ''Anti''-β-elimination reaction catalysed by pectate lyase. R and R’ represent additional galacturonan residues]] |
| − | The PL1 family polysaccharide lyases harness ''anti''-β-elimination chemistry to cleave 1,4-linked | + | The PL1 family polysaccharide lyases ([http://www.cazy.org/PL1.html PL1]) harness ''anti''-β-elimination chemistry to cleave α-(1,4)-linked {{smallcaps|d}}-galacturonan to produce oligosaccharides with an unsaturated hexenuronic acid residue and a new reducing end ([http://www.enzyme-database.org/query.php?ec=4.2.2.2 EC 4.2.2.2]) <cite>Albersheim1962, Edstrom1964a, Edstrom1964b</cite>. Pectin lyases differ from pectate lyases as they are active against a substrate bearing methyl-ester groups at C6 ([http://www.enzyme-database.org/query.php?ec=4.2.2.10 EC 4.2.2.10]). The geometry of the α1,4-linkage facilitates ''anti''-β-elimination about the C4-glycosidic oxygen bond. |
| − | |||
| − | |||
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| Line 40: | Line 39: | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | [[Image:PL1_active_site_crop.jpg|thumb|400px|'''Figure 2.''' Michaelis complex formed using | + | [[Image:PL1_active_site_crop.jpg|thumb|400px|'''Figure 2.''' Michaelis complex formed using ''Bacillus subtilis'' pectate lyase R279A mutant and trigalacturonic acid. The galacturonan residues are labelled according to the subsites they bind: -1, +1, +2. The catalytic arginine is modeled back in the position seen in the native structure and is close to the C5 proton abstracted. The two "catalytic" calcium-ions bound between enzyme and substrate carboxylates that acidify the C5 proton are Ca2 and Ca3.]] |
| − | Acidification of the C5 proton in the polysaccharide family 1 enzymes is by the binding of two “catalytic” calcium-ions which are bound only in the Michaelis complex and not to the free enzyme | + | Acidification of the C5 proton in the polysaccharide family 1 enzymes is by the binding of two “catalytic” calcium-ions which are bound only in the Michaelis complex and not to the free enzyme <cite>Scavetta1999, Herron2000, Seyedarabi2010</cite>. Abstraction of the C5 proton is by the catalytic arginine, the Brønstead base <cite>Scavetta1999, Herron2000, Seyedarabi2010</cite>. The ''Bacillus subtilis'' Michaelis complex is shown in Fig. 2. Protonation of the leaving group is probably by solvent water, though other possibilities have been discussed in the literature. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | Pectate lyases PelC (endo-acting) | + | Pectate lyases PelC (endo-acting) <cite>Yoder1993</cite> and PelE (exo-acting) <cite>Lietzke1994</cite> from ''Erwinia chrysanthemi'', ([{{PDBlink}}2pec 2PEC]) and endo-acting ''Bacillus subtilis'' pectate lyase <cite>Pickersgill1994</cite> ([{{PDBlink}}1bn8 1BN8]). These PL1 structures are remarkable in showing the polypeptide chain folded into a right-handed superhelix comprising three β-strands per turn with turns stacking to form a domain of three parallel β-sheets. The structures showed that parallel β-sheets are stable in the absence of protecting α-helices and revealed remarkable side-chain stacks particularly in the hydrophobic interior. Structures of pectin lyase followed <cite>Mayans1997, Vitali1998</cite>, they are closely similar to pectate lyase have the arginine acting as Brønstead base but also have more hydrophobic active centres suitable for binding methylated pectin. The β-helix fold is conserved in other PL families including [[PL3]] and [[PL9]]. |
== Family Firsts == | == Family Firsts == | ||
| − | ;First description of catalytic activity: Pectin trans-eliminase activity was first demonstrated in 1962 by Albersheim | + | ;First description of catalytic activity: Pectin trans-eliminase activity was first demonstrated in 1962 by Albersheim <cite>Albersheim1962</cite>. |
| − | ;First catalytic base identification: Arginine | + | ;First catalytic base identification: Arginine was identified as the catalytic base from the complex of PelC with pentagalacturonate formed using the inactive mutant in which the arginine was substituted by lysine <cite>Scavetta1999, Herron2000</cite>. The structure showed that the arginine would be in the correct position to abstract the C5 proton. Comparison of PL10 and PL1 Michaelis complexes cemented the role of the arginine as Brønstead base <cite>Charnock2002</cite>. More detailed information on specificity emerged from a more recent study <cite>Seyedarabi2010</cite> which showed the central three subsites bound galacturonsyl-residues, but that the more remote subsites could tolerate methylated galacturonsyl-residues. |
| − | ;First catalytic divalent cation identification: The importance of acidifying the C5 proton by stabilizing the charge on the substrate carboxylate was acknowledged early though the conserved arginine was originally thought to fulfil this function | + | ;First catalytic divalent cation identification: The importance of acidifying the C5 proton by stabilizing the charge on the substrate carboxylate was acknowledged early though the conserved arginine was originally thought to fulfil this function <cite>Pickersgill1994</cite>. It was later shown that two catalytic calcium-ions bound in the Michaelis complex acidify the C5 proton facilitating its abstraction <cite>Scavetta1999, Herron2000</cite>. |
| − | ;First 3-D structures: ''Erwinia chrysanthemi'' | + | ;First 3-D structures: The first structures of PL1s reported were PelC <cite>Yoder1993</cite> and PelE <cite>Lietzke1994</cite> from ''Erwinia chrysanthemi''. The first PL1 structure from ''Bacillus subtilis'' was reported in 1994 <cite>Pickersgill1994</cite>. |
== References == | == References == | ||
| Line 58: | Line 57: | ||
#Edstrom1964b pmid=14235514 | #Edstrom1964b pmid=14235514 | ||
#Gerlt1993 pmid=8218268 | #Gerlt1993 pmid=8218268 | ||
| + | #Scavetta1999 pmid=10368179 | ||
| + | #Herron2000 pmid=10922032 | ||
| + | #Seyedarabi2010 pmid=20000851 | ||
| + | #Yoder1993 pmid=8502994 | ||
| + | #Lietzke1994 pmid=12232373 | ||
| + | #Pickersgill1994 pmid=7634076 | ||
| + | #Mayans1997 pmid=9195887 | ||
| + | #Vitali1998 pmid=9449837 | ||
| + | #Charnock2002 pmid=12221284 | ||
| + | </biblio> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[Category:Polysaccharide Lyase Families|PL001]] | [[Category:Polysaccharide Lyase Families|PL001]] | ||
Latest revision as of 13:16, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Polysaccharide Lyase Family PL1 | |
| 3-D Structure | β-helix |
| Mechanism | β-elimination |
| Charge neutraliser | calcium |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/PL1.html | |
Substrate specificities
The PL1 family polysaccharide lyases (PL1) harness anti-β-elimination chemistry to cleave α-(1,4)-linked d-galacturonan to produce oligosaccharides with an unsaturated hexenuronic acid residue and a new reducing end (EC 4.2.2.2) [1, 2, 3]. Pectin lyases differ from pectate lyases as they are active against a substrate bearing methyl-ester groups at C6 (EC 4.2.2.10). The geometry of the α1,4-linkage facilitates anti-β-elimination about the C4-glycosidic oxygen bond.
Kinetics and Mechanism
The mechanism involves acidification of the C5 proton of the galacturonan residue binding to the +1 subsite of the enzyme, abstraction of this acidified proton, and subsequent leaving group elimination. The leaving group provides a new reducing end and the group from which it was eliminated has an unsaturated C4-C5 bond. The mechanism of proton abstraction from carbon acids is discussed by Gerlt and Gassman [4].
Catalytic Residues
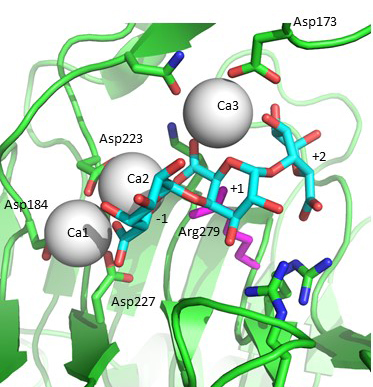
Acidification of the C5 proton in the polysaccharide family 1 enzymes is by the binding of two “catalytic” calcium-ions which are bound only in the Michaelis complex and not to the free enzyme [5, 6, 7]. Abstraction of the C5 proton is by the catalytic arginine, the Brønstead base [5, 6, 7]. The Bacillus subtilis Michaelis complex is shown in Fig. 2. Protonation of the leaving group is probably by solvent water, though other possibilities have been discussed in the literature.
Three-dimensional structures
Pectate lyases PelC (endo-acting) [8] and PelE (exo-acting) [9] from Erwinia chrysanthemi, (2PEC) and endo-acting Bacillus subtilis pectate lyase [10] (1BN8). These PL1 structures are remarkable in showing the polypeptide chain folded into a right-handed superhelix comprising three β-strands per turn with turns stacking to form a domain of three parallel β-sheets. The structures showed that parallel β-sheets are stable in the absence of protecting α-helices and revealed remarkable side-chain stacks particularly in the hydrophobic interior. Structures of pectin lyase followed [11, 12], they are closely similar to pectate lyase have the arginine acting as Brønstead base but also have more hydrophobic active centres suitable for binding methylated pectin. The β-helix fold is conserved in other PL families including PL3 and PL9.
Family Firsts
- First description of catalytic activity
- Pectin trans-eliminase activity was first demonstrated in 1962 by Albersheim [1].
- First catalytic base identification
- Arginine was identified as the catalytic base from the complex of PelC with pentagalacturonate formed using the inactive mutant in which the arginine was substituted by lysine [5, 6]. The structure showed that the arginine would be in the correct position to abstract the C5 proton. Comparison of PL10 and PL1 Michaelis complexes cemented the role of the arginine as Brønstead base [13]. More detailed information on specificity emerged from a more recent study [7] which showed the central three subsites bound galacturonsyl-residues, but that the more remote subsites could tolerate methylated galacturonsyl-residues.
- First catalytic divalent cation identification
- The importance of acidifying the C5 proton by stabilizing the charge on the substrate carboxylate was acknowledged early though the conserved arginine was originally thought to fulfil this function [10]. It was later shown that two catalytic calcium-ions bound in the Michaelis complex acidify the C5 proton facilitating its abstraction [5, 6].
- First 3-D structures
- The first structures of PL1s reported were PelC [8] and PelE [9] from Erwinia chrysanthemi. The first PL1 structure from Bacillus subtilis was reported in 1994 [10].
References
- ALBERSHEIM P and KILLIAS U. (1962). Studies relating to the purification and properties of pectin transeliminase. Arch Biochem Biophys. 1962;97:107-15. DOI:10.1016/0003-9861(62)90050-4 |
- EDSTROM RD and PHAFF HJ. (1964). ELIMINATIVE CLEAVAGE OF PECTIN AND OF OLIGOGALACTURONIDE METHYL ESTERS BY PECTIN TRANS-ELIMINASE. J Biol Chem. 1964;239:2409-15. | Google Books | Open Library
- EDSTROM RD and PHAFF HJ. (1964). PURIFICATION AND CERTAIN PROPERTIES OF PECTIN TRANS-ELIMINASE FROM ASPERGILLUS FONSECAEUS. J Biol Chem. 1964;239:2403-8. | Google Books | Open Library
- Gerlt JA and Gassman PG. (1993). Understanding the rates of certain enzyme-catalyzed reactions: proton abstraction from carbon acids, acyl-transfer reactions, and displacement reactions of phosphodiesters. Biochemistry. 1993;32(45):11943-52. DOI:10.1021/bi00096a001 |
- Scavetta RD, Herron SR, Hotchkiss AT, Kita N, Keen NT, Benen JA, Kester HC, Visser J, and Jurnak F. (1999). Structure of a plant cell wall fragment complexed to pectate lyase C. Plant Cell. 1999;11(6):1081-92. DOI:10.1105/tpc.11.6.1081 |
- Herron SR, Benen JA, Scavetta RD, Visser J, and Jurnak F. (2000). Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc Natl Acad Sci U S A. 2000;97(16):8762-9. DOI:10.1073/pnas.97.16.8762 |
- Seyedarabi A, To TT, Ali S, Hussain S, Fries M, Madsen R, Clausen MH, Teixteira S, Brocklehurst K, and Pickersgill RW. (2010). Structural insights into substrate specificity and the anti beta-elimination mechanism of pectate lyase. Biochemistry. 2010;49(3):539-46. DOI:10.1021/bi901503g |
- Yoder MD, Keen NT, and Jurnak F. (1993). New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260(5113):1503-7. DOI:10.1126/science.8502994 |
- Lietzke SE, Yoder MD, Keen NT, and Jurnak F. (1994). The Three-Dimensional Structure of Pectate Lyase E, a Plant Virulence Factor from Erwinia chrysanthemi. Plant Physiol. 1994;106(3):849-862. DOI:10.1104/pp.106.3.849 |
- Pickersgill R, Jenkins J, Harris G, Nasser W, and Robert-Baudouy J. (1994). The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat Struct Biol. 1994;1(10):717-23. DOI:10.1038/nsb1094-717 |
- Mayans O, Scott M, Connerton I, Gravesen T, Benen J, Visser J, Pickersgill R, and Jenkins J. (1997). Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure. 1997;5(5):677-89. DOI:10.1016/s0969-2126(97)00222-0 |
- Vitali J, Schick B, Kester HC, Visser J, and Jurnak F. (1998). The tree-dimensional structure of aspergillus niger pectin lyase B at 1.7-A resolution. Plant Physiol. 1998;116(1):69-80. DOI:10.1104/pp.116.1.69 |
- Charnock SJ, Brown IE, Turkenburg JP, Black GW, and Davies GJ. (2002). Convergent evolution sheds light on the anti-beta -elimination mechanism common to family 1 and 10 polysaccharide lyases. Proc Natl Acad Sci U S A. 2002;99(19):12067-72. DOI:10.1073/pnas.182431199 |