CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 12"
Harry Brumer (talk | contribs) (added Gilkes1991 ref. for original Cellulase Family H name.) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | * [[Author]]s: | + | * [[Author]]s: [[User:Gerlind Sulzenbacher|Gerlind Sulzenbacher]] and [[User:Mats Sandgren|Mats Sandgren]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Gerlind Sulzenbacher|Gerlind Sulzenbacher]] |
---- | ---- | ||
| Line 28: | Line 28: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | The substrate specificities found among the [[glycoside hydrolases]] of family 12 | + | The substrate specificities found among the [[glycoside hydrolases]] of family 12 include: ''[[endo]]''-β-1,4-glucanase (EC [{{EClink}}3.2.1.4 3.2.1.4]), xyloglucan ''[[endo]]''-hydrolase (EC [{{EClink}}3.2.1.151 3.2.1.151]), and ''[[endo]]''-β-1,3-1,4-glucanase (EC [{{EClink}}3.2.1.73 3.2.1.73]). Xyloglucan ''[[endo]]''-transglycosylase (XET, EC [{{EClink}}2.4.1.207 2.4.1.207]) activity has been observed in a single fungal GH12 member (GenBank [http://www.ncbi.nlm.nih.gov/protein/AAN89225.1 AAN89225.1]) using a XET-specific screen <cite>Nielsen2002</cite>, although this may represent a side activity of a predominant xyloglucan ''[[endo]]''-hydrolase <cite>Gilbert2008 Eklof2010</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | GH12 enzymes are [[retaining]] enzymes, as first shown by NMR studies <cite>Schou1993</cite> on endoglucanase 3 from ''Humicola insolens'' | + | GH12 enzymes are [[retaining]] enzymes, as first shown by NMR studies <cite>Schou1993</cite> on endoglucanase 3 from ''Humicola insolens''. They are believed to follow a classical [[Koshland double-displacement mechanism]] in which a glycosyl-enzyme intermediate is formed and subsequently this intermediate is hydrolysed via an oxocarbenium-ion [[transition state]]. |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | The [[catalytic nucleophile]] and the [[general acid/base]] | + | Families [[GH11]] and GH12 together form [[clan]] GH-C. The [[catalytic nucleophile]] and the [[general acid/base]] residues of family GH12 members (previously known as "family H cellulases" <cite>Gilkes1991 Torronen1993</cite>) was initially predicted by sequence homology to the xylanase members of [[GH11]] <cite>Torronen1993</cite>. Within family [[GH11]] the [[catalytic nucleophile]] was first identified in the ''Bacillus circulans'' [[endo]]-xylanase through trapping of the 2-deoxy-2-fluoroxylobiosyl-enzyme [[intermediate]] and subsequent peptide mapping using LC-MS/MS <cite>Miao1994</cite>. The prediction of the [[catalytic nucleophile]] and the [[general acid/base]] residues of family GH12 enzymes was later confirmed when the first three dimensional structure of a family GH12 enzyme was determined, that of ''Streptomyces lividans'' CelB2 <cite>Sulzenbacher1997</cite>. Subsequently, the [[catalytic nucleophile]] in ''Streptomyces lividans'' CelB2 was assigned as Glu 120 by using the same labeling strategy used for identifying the [[catalytic nucleophile]] of family [[GH11]] <cite>Zechel1998</cite>. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | [[File:gh12_1.png|thumb|300px|right|'''Figure 1: ''Streptomyces lividans'' CelB2 in complex with a 2-deoxy-2-fluorocellotrioside (PDB [{{PDBlink}}2nrl 2NLR]).''' Two distinct species were | + | [[File:gh12_1.png|thumb|300px|right|'''Figure 1: ''Streptomyces lividans'' CelB2 in complex with a 2-deoxy-2-fluorocellotrioside (PDB [{{PDBlink}}2nrl 2NLR]). ''' Two distinct species were modelled in the structure: the glycosyl-enzyme intermediate with the mechanism-based inhibitor covalently linked to the nucleophile Glu120, and the hydrolysis product, 2-deoxy-2-fluorocellotriose. ''Colour coding:'' CelB2 in rainbow colour-ramping from the N-terminus in blue to the C-terminus in red; catalytic residues in magenta; bound oligosaccharide in black]] |
| − | Enzymes from family GH12 adopt a “jelly-roll” fold with two twisted, mostly antiparallel β-sheets stacking on top of each other and enclosing a long substrate-binding groove located on the concave face of the sheet (Fig. 1). A single α-helix packs against the convex surface of the outer β-sheet. GH12 members of bacterial origin feature an additional two-stranded β-sheet, stabilized by a | + | Enzymes from family GH12 adopt a “jelly-roll” fold with two twisted, mostly antiparallel β-sheets stacking on top of each other and enclosing a long substrate-binding groove located on the concave face of the sheet (Fig. 1). A single α-helix packs against the convex surface of the outer β-sheet. GH12 members of bacterial origin feature an additional two-stranded β-sheet, stabilized by a disulfide bridge, located at the rim of the substrate binding groove and providing an additional substrate binding platform as compared to the eukaryotic counterparts. |
| − | The first | + | The first structures for family GH12 were that of ''Streptomyces lividans'' CelB2 in the apo-form <cite>Sulzenbacher1997</cite> and in the form of a glycosyl-enzyme intermediate <cite>Sulzenbacher1999</cite>. These structures confirmed previous predictions, based on hydrophobic cluster analysis <cite>Torronen1993</cite>, that GH12 enzymes would display significant structural similarity with family [[GH11]] xylanases and gave rise to the creation of [[clan]] GH-C, which so far comprises only these two families. The first structure of a fungal GH12 enzyme was determined shortly thereafter, for ''Trichoderma reesei'' (formerly ''Hypocrea jecorina'') Cel12A <cite>Sandgren2001</cite>. To date structural models, both in the apo-form and in complex with oligosaccharides, are available for sixteen family members. |
| − | + | The substrate-binding grooves of family GH12 enzymes harbour six binding sites, from -4 to +2, lined by numerous aromatic residues providing binding platforms for the individual pyranose units. This is consistent with the endoglucanase activity of these enzymes, exhibiting low catalytic efficiency towards short chain substrates <cite>Karlsson2002</cite>. The [[catalytic nucleophile]] Glu120 (''S. lividans'' CelB2 numbering) and the [[general acid/base]] Glu203 are in close proximity of an invariant aspartate, which might have an important role in catalysis. This catalytic triad is reminiscent of the catalytic centre of family [[GH7]] enzymes. GH12 family members, like those of family [[GH7]], have the acid/base residue situated 'syn' to the pyranoside 0-5-C-1 bond, and are thus classified as [[Syn/anti_lateral_protonation|syn-protonators]]. | |
[[File:gh12_2.png|thumb|300px|right|'''Figure 2: Xyloglucanase XG12 from ''Bacillus licheniformis'' in complex with xyloglucan (PDB [{{PDBlink}}}2jen 2JEN]).''' α-1,6-xylose substitutions are present at subsites -3, -2, +1 and +2.]] | [[File:gh12_2.png|thumb|300px|right|'''Figure 2: Xyloglucanase XG12 from ''Bacillus licheniformis'' in complex with xyloglucan (PDB [{{PDBlink}}}2jen 2JEN]).''' α-1,6-xylose substitutions are present at subsites -3, -2, +1 and +2.]] | ||
| − | + | Structural and function studies of several xyloglucan-specific enzymes of the family, including xyloglucanase XG12 from ''Bacillus licheniformis'' <cite>Gloster2007</cite> and the xyloglucan specific endo-β-1,4-glucanase XEG1 from ''Aspergillus aculeatus KSM 50'' <cite>Yoshizawa2012</cite>, have shown that α-1,6-xylose substitutions can be tolerated at subsites -3, -2, +1 and +2 (Fig. 2). Certain GH12 enzymes exhibit β-1,3-1,4-glucanase activity and the crystal structure of ''Humicola grisea'' Cel12A in complex with a mixed linkage oligosaccharide arising from a transglycosylation reaction revealed that the β-1,3-linkage can be accommodated between subsites -3 and -2 <cite>Sandgren2004</cite>. It has been speculated that a long loop crossing the substrate-binding groove at its reducing end, known as the “cord” and conserved in all [[clan]] GH-C members, might undergo conformational changes upon substrate binding <cite>Torronen1994</cite>, but the available GH12 complex structures support the view that the role of this loop is to deliver residues for substrate binding. | |
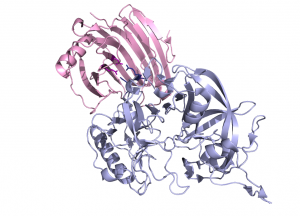
| − | The crystal structure of Aspergillus aculeatus XEG1 in complex with the glycoside hydrolase inhibitory protein from carrot | + | The crystal structure of ''Aspergillus aculeatus'' XEG1 in complex with the glycoside hydrolase inhibitory protein (GHIP) from carrot, EDGP, reveals a mode of inhibition distinct from that observed previously for the inhibition of a family [[GH11]] xylanase by a wheat protein <cite>Payan2004</cite>. In the XEG1-GHIP complex the inhibitor mimics a xyloglucan substrate and inserts two conserved arginine residues into the active site, which establish salt bridges with the carboxylate groups of the catalytic centre (Fig. 3) <cite>Yoshizawa2012</cite>). |
[[File:gh12_3.png|thumb|300px|right|'''Figure 3: Inhibition of ''Aspergillus aculeatus'' XEG1 by the glycoside hydrolase inhibitory protein from carrot. EDGP (PDB [{{PDBlink}}3vlb 3VLB]).''' Two conserved arginine residues of the inhibitor make salt bridges with the catalytic residues of XEG1. ''Colour coding:'' XEG1 in pink with carboxylate groups of the catalytic centre in purple; EDGP in lightblue with the conserved arginine residues in deepblue.]] | [[File:gh12_3.png|thumb|300px|right|'''Figure 3: Inhibition of ''Aspergillus aculeatus'' XEG1 by the glycoside hydrolase inhibitory protein from carrot. EDGP (PDB [{{PDBlink}}3vlb 3VLB]).''' Two conserved arginine residues of the inhibitor make salt bridges with the catalytic residues of XEG1. ''Colour coding:'' XEG1 in pink with carboxylate groups of the catalytic centre in purple; EDGP in lightblue with the conserved arginine residues in deepblue.]] | ||
| − | Due to | + | Due to commercial interest in their bioprocessing applications, GH12 endo-β-1,4-glucanases from ''Humicola grisea'' <cite>Sandgren2003a</cite>, ''Trichoderma reesei'' <cite>Sandgren2003a Sandgren2003b</cite>, ''Rhodothermus marinus'' <cite>Kapoor2008</cite>, ''Pyrococcus furiosus'' <cite>Kim2012</cite> and ''Thermotoga maritima'' <cite>Cheng2012</cite> have been extensively engineered and the structural studies of the resulting variants have provided valuable insight into structural features governing enzyme activity and stability. |
== Family Firsts == | == Family Firsts == | ||
| − | ;First | + | ;First stereochemistry determination: ''Humicola insolens'' endoglucanase 3 by <sup>1</sup>H NMR <cite>Schou1993</cite>. |
| − | ;First [[catalytic nucleophile]] identification: The catalytic nucleophile was first identified in ''Streptomyces lividans'' CelB2 by trapping of a glycosyl-enzyme intermediate | + | ;First [[catalytic nucleophile]] identification: The catalytic nucleophile was first identified in ''Streptomyces lividans'' CelB2 by trapping of a glycosyl-enzyme intermediate and X-ray structure determination <cite>Sulzenbacher1999</cite> and peptide mapping using LC-MS/MS <cite>Zechel1998</cite>. |
| − | ;First [[general acid/base]] residue identification: | + | ;First [[general acid/base]] residue identification: Inferred from homology with family [[GH11]] enzymes. |
| − | ;First 3-D structure: The crystal structure of ''Streptomyces lividans'' CelB2 <cite>Sulzenbacher1997</cite> | + | ;First 3-D structure: The crystal structure of ''Streptomyces lividans'' CelB2 <cite>Sulzenbacher1997</cite>. |
== References == | == References == | ||
Latest revision as of 13:18, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH12 | |
| Clan | GH-C |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH12.html | |
Substrate specificities
The substrate specificities found among the glycoside hydrolases of family 12 include: endo-β-1,4-glucanase (EC 3.2.1.4), xyloglucan endo-hydrolase (EC 3.2.1.151), and endo-β-1,3-1,4-glucanase (EC 3.2.1.73). Xyloglucan endo-transglycosylase (XET, EC 2.4.1.207) activity has been observed in a single fungal GH12 member (GenBank AAN89225.1) using a XET-specific screen [1], although this may represent a side activity of a predominant xyloglucan endo-hydrolase [2, 3].
Kinetics and Mechanism
GH12 enzymes are retaining enzymes, as first shown by NMR studies [4] on endoglucanase 3 from Humicola insolens. They are believed to follow a classical Koshland double-displacement mechanism in which a glycosyl-enzyme intermediate is formed and subsequently this intermediate is hydrolysed via an oxocarbenium-ion transition state.
Catalytic Residues
Families GH11 and GH12 together form clan GH-C. The catalytic nucleophile and the general acid/base residues of family GH12 members (previously known as "family H cellulases" [5, 6]) was initially predicted by sequence homology to the xylanase members of GH11 [6]. Within family GH11 the catalytic nucleophile was first identified in the Bacillus circulans endo-xylanase through trapping of the 2-deoxy-2-fluoroxylobiosyl-enzyme intermediate and subsequent peptide mapping using LC-MS/MS [7]. The prediction of the catalytic nucleophile and the general acid/base residues of family GH12 enzymes was later confirmed when the first three dimensional structure of a family GH12 enzyme was determined, that of Streptomyces lividans CelB2 [8]. Subsequently, the catalytic nucleophile in Streptomyces lividans CelB2 was assigned as Glu 120 by using the same labeling strategy used for identifying the catalytic nucleophile of family GH11 [9].
Three-dimensional structures
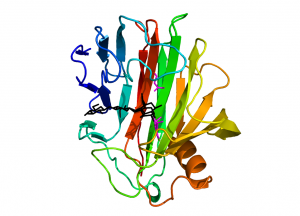
Enzymes from family GH12 adopt a “jelly-roll” fold with two twisted, mostly antiparallel β-sheets stacking on top of each other and enclosing a long substrate-binding groove located on the concave face of the sheet (Fig. 1). A single α-helix packs against the convex surface of the outer β-sheet. GH12 members of bacterial origin feature an additional two-stranded β-sheet, stabilized by a disulfide bridge, located at the rim of the substrate binding groove and providing an additional substrate binding platform as compared to the eukaryotic counterparts.
The first structures for family GH12 were that of Streptomyces lividans CelB2 in the apo-form [8] and in the form of a glycosyl-enzyme intermediate [10]. These structures confirmed previous predictions, based on hydrophobic cluster analysis [6], that GH12 enzymes would display significant structural similarity with family GH11 xylanases and gave rise to the creation of clan GH-C, which so far comprises only these two families. The first structure of a fungal GH12 enzyme was determined shortly thereafter, for Trichoderma reesei (formerly Hypocrea jecorina) Cel12A [11]. To date structural models, both in the apo-form and in complex with oligosaccharides, are available for sixteen family members.
The substrate-binding grooves of family GH12 enzymes harbour six binding sites, from -4 to +2, lined by numerous aromatic residues providing binding platforms for the individual pyranose units. This is consistent with the endoglucanase activity of these enzymes, exhibiting low catalytic efficiency towards short chain substrates [12]. The catalytic nucleophile Glu120 (S. lividans CelB2 numbering) and the general acid/base Glu203 are in close proximity of an invariant aspartate, which might have an important role in catalysis. This catalytic triad is reminiscent of the catalytic centre of family GH7 enzymes. GH12 family members, like those of family GH7, have the acid/base residue situated 'syn' to the pyranoside 0-5-C-1 bond, and are thus classified as syn-protonators.
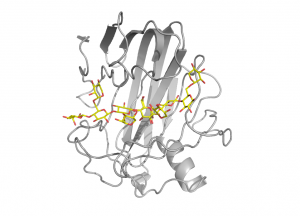
Structural and function studies of several xyloglucan-specific enzymes of the family, including xyloglucanase XG12 from Bacillus licheniformis [13] and the xyloglucan specific endo-β-1,4-glucanase XEG1 from Aspergillus aculeatus KSM 50 [14], have shown that α-1,6-xylose substitutions can be tolerated at subsites -3, -2, +1 and +2 (Fig. 2). Certain GH12 enzymes exhibit β-1,3-1,4-glucanase activity and the crystal structure of Humicola grisea Cel12A in complex with a mixed linkage oligosaccharide arising from a transglycosylation reaction revealed that the β-1,3-linkage can be accommodated between subsites -3 and -2 [15]. It has been speculated that a long loop crossing the substrate-binding groove at its reducing end, known as the “cord” and conserved in all clan GH-C members, might undergo conformational changes upon substrate binding [16], but the available GH12 complex structures support the view that the role of this loop is to deliver residues for substrate binding.
The crystal structure of Aspergillus aculeatus XEG1 in complex with the glycoside hydrolase inhibitory protein (GHIP) from carrot, EDGP, reveals a mode of inhibition distinct from that observed previously for the inhibition of a family GH11 xylanase by a wheat protein [17]. In the XEG1-GHIP complex the inhibitor mimics a xyloglucan substrate and inserts two conserved arginine residues into the active site, which establish salt bridges with the carboxylate groups of the catalytic centre (Fig. 3) [14]).
Due to commercial interest in their bioprocessing applications, GH12 endo-β-1,4-glucanases from Humicola grisea [18], Trichoderma reesei [18, 19], Rhodothermus marinus [20], Pyrococcus furiosus [21] and Thermotoga maritima [22] have been extensively engineered and the structural studies of the resulting variants have provided valuable insight into structural features governing enzyme activity and stability.
Family Firsts
- First stereochemistry determination
- Humicola insolens endoglucanase 3 by 1H NMR [4].
- First catalytic nucleophile identification
- The catalytic nucleophile was first identified in Streptomyces lividans CelB2 by trapping of a glycosyl-enzyme intermediate and X-ray structure determination [10] and peptide mapping using LC-MS/MS [9].
- First general acid/base residue identification
- Inferred from homology with family GH11 enzymes.
- First 3-D structure
- The crystal structure of Streptomyces lividans CelB2 [8].
References
-
Nielsen, RI. (2002) Microbial xyloglucan endotransglycosylase (XET) Patent US6448056.
- Gilbert HJ, Stålbrand H, and Brumer H. (2008). How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol. 2008;11(3):338-48. DOI:10.1016/j.pbi.2008.03.004 |
- Eklöf JM and Brumer H. (2010). The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010;153(2):456-66. DOI:10.1104/pp.110.156844 |
- Schou C, Rasmussen G, Kaltoft MB, Henrissat B, and Schülein M. (1993). Stereochemistry, specificity and kinetics of the hydrolysis of reduced cellodextrins by nine cellulases. Eur J Biochem. 1993;217(3):947-53. DOI:10.1111/j.1432-1033.1993.tb18325.x |
- Gilkes NR, Henrissat B, Kilburn DG, Miller RC Jr, and Warren RA. (1991). Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55(2):303-15. DOI:10.1128/mr.55.2.303-315.1991 |
- Törrönen A, Kubicek CP, and Henrissat B. (1993). Amino acid sequence similarities between low molecular weight endo-1,4-beta-xylanases and family H cellulases revealed by clustering analysis. FEBS Lett. 1993;321(2-3):135-9. DOI:10.1016/0014-5793(93)80094-b |
- Miao S, Ziser L, Aebersold R, and Withers SG. (1994). Identification of glutamic acid 78 as the active site nucleophile in Bacillus subtilis xylanase using electrospray tandem mass spectrometry. Biochemistry. 1994;33(23):7027-32. DOI:10.1021/bi00189a002 |
- Sulzenbacher G, Shareck F, Morosoli R, Dupont C, and Davies GJ. (1997). The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution. Biochemistry. 1997;36(51):16032-9. DOI:10.1021/bi972407v |
- Zechel DL, He S, Dupont C, and Withers SG. (1998). Identification of Glu-120 as the catalytic nucleophile in Streptomyces lividans endoglucanase celB. Biochem J. 1998;336 ( Pt 1)(Pt 1):139-45. DOI:10.1042/bj3360139 |
- Sulzenbacher G, Mackenzie LF, Wilson KS, Withers SG, Dupont C, and Davies GJ. (1999). The crystal structure of a 2-fluorocellotriosyl complex of the Streptomyces lividans endoglucanase CelB2 at 1.2 A resolution. Biochemistry. 1999;38(15):4826-33. DOI:10.1021/bi982648i |
- Sandgren M, Shaw A, Ropp TH, Wu S, Bott R, Cameron AD, Ståhlberg J, Mitchinson C, and Jones TA. (2001). The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 A resolution. J Mol Biol. 2001;308(2):295-310. DOI:10.1006/jmbi.2001.4583 |
- Karlsson J, Siika-aho M, Tenkanen M, and Tjerneld F. (2002). Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J Biotechnol. 2002;99(1):63-78. DOI:10.1016/s0168-1656(02)00156-6 |
- Gloster TM, Ibatullin FM, Macauley K, Eklöf JM, Roberts S, Turkenburg JP, Bjørnvad ME, Jørgensen PL, Danielsen S, Johansen KS, Borchert TV, Wilson KS, Brumer H, and Davies GJ. (2007). Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J Biol Chem. 2007;282(26):19177-89. DOI:10.1074/jbc.M700224200 |
- Yoshizawa T, Shimizu T, Hirano H, Sato M, and Hashimoto H. (2012). Structural basis for inhibition of xyloglucan-specific endo-β-1,4-glucanase (XEG) by XEG-protein inhibitor. J Biol Chem. 2012;287(22):18710-6. DOI:10.1074/jbc.M112.350520 |
- Sandgren M, Berglund GI, Shaw A, Ståhlberg J, Kenne L, Desmet T, and Mitchinson C. (2004). Crystal complex structures reveal how substrate is bound in the -4 to the +2 binding sites of Humicola grisea Cel12A. J Mol Biol. 2004;342(5):1505-17. DOI:10.1016/j.jmb.2004.07.098 |
- Törrönen A, Harkki A, and Rouvinen J. (1994). Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO J. 1994;13(11):2493-501. DOI:10.1002/j.1460-2075.1994.tb06536.x |
- Payan F, Leone P, Porciero S, Furniss C, Tahir T, Williamson G, Durand A, Manzanares P, Gilbert HJ, Juge N, and Roussel A. (2004). The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases. J Biol Chem. 2004;279(34):36029-37. DOI:10.1074/jbc.M404225200 |
- Sandgren M, Gualfetti PJ, Paech C, Paech S, Shaw A, Gross LS, Saldajeno M, Berglund GI, Jones TA, and Mitchinson C. (2003). The Humicola grisea Cel12A enzyme structure at 1.2 A resolution and the impact of its free cysteine residues on thermal stability. Protein Sci. 2003;12(12):2782-93. DOI:10.1110/ps.03220403 |
- Sandgren M, Gualfetti PJ, Shaw A, Gross LS, Saldajeno M, Day AG, Jones TA, and Mitchinson C. (2003). Comparison of family 12 glycoside hydrolases and recruited substitutions important for thermal stability. Protein Sci. 2003;12(4):848-60. DOI:10.1110/ps.0237703 |
- Kapoor D, Kumar V, Chandrayan SK, Ahmed S, Sharma S, Datt M, Singh B, Karthikeyan S, and Guptasarma P. (2008). Replacement of the active surface of a thermophile protein by that of a homologous mesophile protein through structure-guided 'protein surface grafting'. Biochim Biophys Acta. 2008;1784(11):1771-6. DOI:10.1016/j.bbapap.2008.05.007 |
- Kim HW, Kataoka M, and Ishikawa K. (2012). Atomic resolution of the crystal structure of the hyperthermophilic family 12 endocellulase and stabilizing role of the DxDxDG calcium-binding motif in Pyrococcus furiosus. FEBS Lett. 2012;586(7):1009-13. DOI:10.1016/j.febslet.2012.02.029 |
- Cheng YS, Ko TP, Huang JW, Wu TH, Lin CY, Luo W, Li Q, Ma Y, Huang CH, Wang AH, Liu JR, and Guo RT. (2012). Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop. Appl Microbiol Biotechnol. 2012;95(3):661-9. DOI:10.1007/s00253-011-3791-4 |