CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 72"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (16 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | * [[Author]]s: | + | * [[Author]]s: [[User:Ramon Hurtado-Guerrero|Ramon Hurtado-Guerrero]] and [[User:Thierry Fontaine|Thierry Fontaine]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Bernard Henrissat|Bernard Henrissat]] |
---- | ---- | ||
| Line 28: | Line 28: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | + | [[Glycoside hydrolase]] family GH72 is formed exclusively of [[transglycosylases]] of the fungal kingdom. Their activity was first characterized in ''Aspergillus fumigatus'' <cite>Hartland1996</cite> and yeasts <cite>Mouyna2000 Carotti2004 deMedina-Redondo2008</cite>. GH72 transglycosidases are GPI-anchored plasma membrane enzymes that elongate and remodel the 1,3-β-glucan of the cell wall <cite>Mouyna2000a Mouyna2005 Gastebois2010 deMedina-Redondo2008 deMedina-Redondo2010 Ragni2007a</cite>. The catalytic domain is located in the external part of the plasma membrane. Two sub-families have been described for GH72 family members depending on the presence or absence of a C-terminal cysteine rich domain (carbohydrate binding domain, [[CBM43]]) in addition to the catalytic domain <cite>Ragni2007b</cite>. | |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | + | Catalysis by GH72 family enzymes occurs through a [[classical Koshland retaining mechanism]], which leads to net retention of the β-anomeric configuration of the final product. Enzymatic kinetics were determined by HPLC and showed that these enzymes are [[transglycosylases]] rather than [[glycoside hydrolases]]. Fungal GH72 enzymes internally cleave a 1,3-β-glucan molecule and form a glycosyl enzyme which reacts with the non-reducing end of a second β-1,3-glucan molecule, forming a new β-1,3-glucosidic linkage, resulting in the truncation of the first chain and elongation of the second. The minimum size of the donor and acceptor substrates so far described are laminaridecaose and laminaripentaose, respectively <cite>Hartland1996 Mazan2011</cite>. | |
| − | Despite that the overall mechanisms of [[glycoside hydrolases|hydrolysis]] and [[transglycosylases|transglycosylation]] are well known, it is still unclear how [[transglycosylases]] limit or prevent hydrolysis in aqueous media, where the concentration of water is 55 M. By structural studies with different laminarioligosaccharides and enzymatic activity assays, a “base occlusion mechanism”, in which the acceptor sugar blocks the entrance of water molecules, was proposed to explain this phenomenon <cite>Hurtado-Guerrero2009</cite>. | + | Despite the fact that the overall mechanisms of [[glycoside hydrolases|hydrolysis]] and [[transglycosylases|transglycosylation]] are well known, it is still unclear how [[transglycosylases]] limit or prevent hydrolysis in aqueous media, where the concentration of water is 55 M. By structural studies with different laminarioligosaccharides and enzymatic activity assays, a “base occlusion mechanism”, in which the acceptor sugar blocks the entrance of water molecules, was proposed to explain this phenomenon <cite>Hurtado-Guerrero2009</cite>. |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | Multiple sequence alignments have highlighted conserved amino acid between GH72 family members <cite>Mouyna2000b</cite>. Hydrophobic cluster analysis allowed | + | Multiple sequence alignments have highlighted conserved amino acid between GH72 family members <cite>Mouyna2000b</cite>. Hydrophobic cluster analysis allowed identification of two highly conserved glutamate residues in the region comparable to the C-terminal end of strands β-4 and β-7 of ''Clostridium cellulolyticum'' endoglucanase A (a [[GH5]] member) <cite>Mouyna2000</cite>. Site-direct mutagenesis of these two glutamate residues in ''A. fumigatus'' Gel1p and ''S. cerevisiae'' Gas1p have shown their essentiality for the transglycosidase activity <cite>Mouyna2000b Carotti2004</cite> and support the assignment of these residues as the acid-base and nucleophilic residues (Glu-160 and Glu-261, respectively, of Gel1p from ''C. albicans''). The identity of these residues was further confirmed by the resolution of the crystal structure of ''S. cerevisiae'' Gas2 (ScGas2) (see below) <cite>Hurtado-Guerrero2009</cite>. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The | + | [[File:Gas2FINAL.jpg|thumb|300px|right|'''Figure 1.''' Crystal structure of ''Sc''Gas2 ([{{PDBlink}}2w62 PDB ID 2w62]).]]The first three-dimensional structures available for a GH72 member were that of ''S. cerevisiae'' ScGas2 in free form ([{{PDBlink}}2w61 PDB ID 2w61]) and in complex with carbohydrates ([{{PDBlink}}2w62 PDB ID 2w62], [{{PDBlink}}2w63 PDB ID 2w63]) (Figure 1). The enzyme folds as a (beta/alpha)<sub>8</sub> barrel similar to that prevailing in other families constituting Clan GH-A <cite>Hurtado-Guerrero2009</cite>. The full length enzyme also harbors a [[CBM43]] module at the C-terminus. The crystal structure also showed that both domains share extensive contacts <cite>Hurtado-Guerrero2009</cite>. |
== Family Firsts == | == Family Firsts == | ||
;First stereochemistry determination: | ;First stereochemistry determination: | ||
| − | β-1,3- | + | β-1,3-glucanosyltransglycosylase (Gel1p) from ''Aspergillus fumigatus'' <cite>Hartland1996</cite>. |
;First catalytic nucleophile identification: | ;First catalytic nucleophile identification: | ||
| − | Shown in the β-1,3- | + | Shown in the β-1,3-glucanosyltransglycosylase (Gel1p) from ''Aspergillus fumigatus'' <cite>Mouyna2000b</cite>. |
;First general acid/base residue identification: | ;First general acid/base residue identification: | ||
| − | Shown in the β-1,3- | + | Shown in the β-1,3-glucanosyltransglycosylase (Gel1p) from ''Aspergillus fumigatus'' <cite>Mouyna2000b</cite>. |
| − | ;First 3-D structure: ScGas2 crystal structure <cite>Hurtado-Guerrero2009</cite> | + | ;First 3-D structure: |
| + | ScGas2 crystal structure <cite>Hurtado-Guerrero2009</cite>. | ||
== References == | == References == | ||
Latest revision as of 13:19, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH72 | |
| Clan | none, (βα)8 fold |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH72.html | |
Substrate specificities
Glycoside hydrolase family GH72 is formed exclusively of transglycosylases of the fungal kingdom. Their activity was first characterized in Aspergillus fumigatus [1] and yeasts [2, 3, 4]. GH72 transglycosidases are GPI-anchored plasma membrane enzymes that elongate and remodel the 1,3-β-glucan of the cell wall [4, 5, 6, 7, 8, 9]. The catalytic domain is located in the external part of the plasma membrane. Two sub-families have been described for GH72 family members depending on the presence or absence of a C-terminal cysteine rich domain (carbohydrate binding domain, CBM43) in addition to the catalytic domain [10].
Kinetics and Mechanism
Catalysis by GH72 family enzymes occurs through a classical Koshland retaining mechanism, which leads to net retention of the β-anomeric configuration of the final product. Enzymatic kinetics were determined by HPLC and showed that these enzymes are transglycosylases rather than glycoside hydrolases. Fungal GH72 enzymes internally cleave a 1,3-β-glucan molecule and form a glycosyl enzyme which reacts with the non-reducing end of a second β-1,3-glucan molecule, forming a new β-1,3-glucosidic linkage, resulting in the truncation of the first chain and elongation of the second. The minimum size of the donor and acceptor substrates so far described are laminaridecaose and laminaripentaose, respectively [1, 11]. Despite the fact that the overall mechanisms of hydrolysis and transglycosylation are well known, it is still unclear how transglycosylases limit or prevent hydrolysis in aqueous media, where the concentration of water is 55 M. By structural studies with different laminarioligosaccharides and enzymatic activity assays, a “base occlusion mechanism”, in which the acceptor sugar blocks the entrance of water molecules, was proposed to explain this phenomenon [12].
Catalytic Residues
Multiple sequence alignments have highlighted conserved amino acid between GH72 family members [13]. Hydrophobic cluster analysis allowed identification of two highly conserved glutamate residues in the region comparable to the C-terminal end of strands β-4 and β-7 of Clostridium cellulolyticum endoglucanase A (a GH5 member) [2]. Site-direct mutagenesis of these two glutamate residues in A. fumigatus Gel1p and S. cerevisiae Gas1p have shown their essentiality for the transglycosidase activity [3, 13] and support the assignment of these residues as the acid-base and nucleophilic residues (Glu-160 and Glu-261, respectively, of Gel1p from C. albicans). The identity of these residues was further confirmed by the resolution of the crystal structure of S. cerevisiae Gas2 (ScGas2) (see below) [12].
Three-dimensional structures
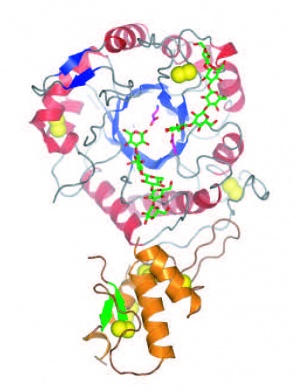
The first three-dimensional structures available for a GH72 member were that of S. cerevisiae ScGas2 in free form (PDB ID 2w61) and in complex with carbohydrates (PDB ID 2w62, PDB ID 2w63) (Figure 1). The enzyme folds as a (beta/alpha)8 barrel similar to that prevailing in other families constituting Clan GH-A [12]. The full length enzyme also harbors a CBM43 module at the C-terminus. The crystal structure also showed that both domains share extensive contacts [12].
Family Firsts
- First stereochemistry determination
β-1,3-glucanosyltransglycosylase (Gel1p) from Aspergillus fumigatus [1].
- First catalytic nucleophile identification
Shown in the β-1,3-glucanosyltransglycosylase (Gel1p) from Aspergillus fumigatus [13].
- First general acid/base residue identification
Shown in the β-1,3-glucanosyltransglycosylase (Gel1p) from Aspergillus fumigatus [13].
- First 3-D structure
ScGas2 crystal structure [12].
References
Error fetching PMID 10809732:
Error fetching PMID 15355340:
Error fetching PMID 18410286:
Error fetching PMID 10809732:
Error fetching PMID 15916615:
Error fetching PMID 20543062:
Error fetching PMID 21124977:
Error fetching PMID 17189486:
Error fetching PMID 17397106:
Error fetching PMID 21651500:
Error fetching PMID 10769178:
- Error fetching PMID 8900166:
- Error fetching PMID 10809732:
- Error fetching PMID 15355340:
- Error fetching PMID 18410286:
- Error fetching PMID 10809732:
- Error fetching PMID 15916615:
- Error fetching PMID 20543062:
- Error fetching PMID 21124977:
- Error fetching PMID 17189486:
- Error fetching PMID 17397106:
- Error fetching PMID 21651500:
- Hurtado-Guerrero R, Schüttelkopf AW, Mouyna I, Ibrahim AF, Shepherd S, Fontaine T, Latgé JP, and van Aalten DM. (2009). Molecular mechanisms of yeast cell wall glucan remodeling. J Biol Chem. 2009;284(13):8461-9. DOI:10.1074/jbc.M807990200 |
- Error fetching PMID 10769178: