CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 84"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (9 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | * [[Author]]: | + | * [[Author]]: [[User:Ian Greig|Ian Greig]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:David Vocadlo|David Vocadlo]] |
---- | ---- | ||
| Line 30: | Line 30: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | GH84 | + | [[Glycoside hydrolases]] of GH84 include ''β''-''N''-acetylglucosaminidases but members have often also been annotated as ''β''-''N''-acetylhyaluronidases. This second annotation arises from the initial cloning of the enzyme and its early sequence analysis, which suggested sequence similarity to ''β''-''N''-acetylhyaluronidases <cite>Heckel98</cite>. Biochemical analysis of a GH84 enzyme annotated in as a hyaluronidase, however, revealed that it had no such activity <cite>Black2006</cite> and structural studies of other homologues have suggested the active site would be unable to process polymeric hyaluronan. Humans possess only one enzyme belonging to GH84, first defined by its unique biochemical properties including activity at neutral pH and selectivity for ''N''-acetylglucosamine residues. The human GH84 enzyme was labelled as HexC to distinguish it from the human [[GH20]] lysosomal enzymes HexA and HexB. HexC was first cloned from a meningoma library using autologous serum and defined as meningoma expressed antigen 5 (MGEA5) <cite>Heckel98</cite>. It was later cloned and biochemically characterized as ''O''-GlcNAcase, a nuclear and cytoplasmic enzyme targeting glycoprotein substrates modified by ''β''-linked GlcNAc residues linked to serine and threonine residues <cite>Gao01 Dong94</cite>. A truncated, nuclear-localized isoform of human ''O''-GlcNAcase lacking a putative C-terminal histone acetyl transferase domain retains similar kinetic properties (aside from a much lower activity) and inhibition patterns as the full-length cytosolic isoform and is consistent with hexosaminidase activity residing in the ''N''-terminal domain, which shares similarity with other GH84 enzymes <cite>DJV2009Trunc</cite>. In contrast to the ''β''-hexosaminidases of [[GH20]] a relaxed specificity for substitutions of the ''N''-acyl group is observed with residues significantly more bulky than the ''N''-acyl group being tolerated <cite>DJV2005</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | Members of GH84 use a catalytic mechanism of [[neighboring group participation]], this originally being established through the use of free-energy relationship-based studies of human ''O''-GlcNAcase <cite>DJV2005</cite>. More recent studies of | + | Members of GH84 are [[retaining]] enzymes that use a catalytic mechanism of [[neighboring group participation]], this originally being established through the use of free-energy relationship-based studies of human ''O''-GlcNAcase <cite>DJV2005</cite>. More recent studies of the human enzyme have investigated variations in rates of reaction (''V/K'') with both nucleophile and leaving group structures <cite>DJV2009</cite>. For substrates possessing the naturally-occurring acetyl nucleophile a pre-chemical step is rate-determining on ''V/K'' for leaving groups with a p''K''<sub>a</sub> below 11 (with the chemical step rate-determining for substrates with higher p''K''<sub>a</sub> values). Studies of substrates possessing fluoroacetyl nucleophiles highlighted that a dissociative (D<sub>N</sub>*A<sub>N</sub>) <cite>Guthrie89</cite> mechanism involving [[general acid]] catalysis operates for the hydrolysis of substrates possessing leaving groups with a p''K''<sub>a</sub> greater than approximately 7; a concerted (A<sub>N</sub>D<sub>N</sub>) <cite>Guthrie89</cite> mechanism, not employing general acid catalysis was found for substrates possessing leaving groups with lower p''K''<sub>a</sub> values (consistent with prior studies showing hydrolysis of 1-''S''-glucosaminides <cite>DJV2005Thio</cite>). |
| − | |||
| − | |||
| + | Numerous carbohydrate and carbohydrate like-scaffolds have been reported as potent inhibitors of GH84 enzymes. These include "NAG-thiazolines" <cite>DJV2005</cite>, PUGNAc (''O''-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino-''N''-phenylcarbamate) <cite>Stubbs06</cite>, GlcNAcstatins <cite>Dorf2006 Vasella2006</cite>, 6-''epi''-valeinamines <cite>Stick2007</cite>, and 6-acetamido-6-deoxy-castanospermine <cite>Mac2010</cite>. The greater tolerance of the ''N''-acyl binding pockets of GH84 enzymes for bulky sidechains compared to those of [[GH20]] enzymes has enabled the development of inhibitors that are not only highly potent but also highly selective in their inhibition of human nucleocytoplasmic ''O''-GlcNAcase (GH84) over human lysosomal ''β''-hexosaminidases <cite>DJV2005 Whitworth2007 Dorf2010</cite>. Many of these families of inhibitors possess structural characteristics reminiscent of the [[oxocarbenium ion]]-like transition states of glycosyl group transfer and, as such may loosely be termed '[[transition state]] analogues'. An analysis of NAG-thiazoline- and PUGNAc-derived inhibitors of human ''O''-GlcNAcase has shown that only the NAG-thiazolines position the inhibitors and their ''N''-acyl side-chains within the hydrophobic binding pocket in a manner consistent with the species found along the reaction coordinate <cite>Whitworth2007</cite>. As such NAG-thiazoline inhibitors may be termed 'Bartlett-type' (free-energy relationship-based) <cite>Bartlett1997</cite> [[transition state]] analogues. Significantly streptozotocin, a widely used diabetogenic compound whose toxicity towards pancreatic β-cells was hypothesized to arise from its ability to act as an inhibitor of human ''O''-GlcNAcase, neither binds covalently to GH84 enzymes <cite>DJV2005 DvA2008 GJD2009</cite>, nor is a particularly potent inhibitor of human ''O''-GlcNAcase <cite>DJV2005 Kudlow2001 Sato1999</cite>. | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | + | Studies of two mutants of human ''O''-GlcNAcase established that adjacent aspartate residues, Asp174 and Asp175, act as critical components of the catalytic machinery of this enzyme <cite>DJV2006</cite>. | |
| − | Studies of two mutants of human ''O''-GlcNAcase established that adjacent aspartate residues, Asp174 and Asp175, act as critical components of the catalytic machinery of this enzyme | ||
| − | The mutant Asp175Ala displayed marked reductions in activity (''V'' and (''V/K'')) towards aryl ''N''-acetylglucosaminides possessing poor leaving groups with smaller reductions being observed for both ''O''-aryl and ''S''-aryl ''N''-acetylglucosaminides substrates possessing better leaving groups. Exogenous azide was found to partially rescue the activity of human ''O''-GlcNAcase towards 3,4-dinitrophenylglucosaminide. These results clearly identified Asp175 as the general acid catalyst. | + | The mutant Asp175Ala displayed marked reductions in activity (''V'' and (''V/K'')) towards aryl ''N''-acetylglucosaminides possessing poor leaving groups with smaller reductions being observed for both ''O''-aryl and ''S''-aryl ''N''-acetylglucosaminides substrates possessing better leaving groups. Exogenous azide was found to partially rescue the activity of human ''O''-GlcNAcase towards 3,4-dinitrophenylglucosaminide. These results clearly identified Asp175 as the [[general acid]] catalyst. |
The mutant Asp174Ala showed significantly decreased activity towards ''O''-aryl ''N''-acetylglucosaminides possessing either good or poor leaving groups and it was argued that this is consistent with its role as a residue responsible for the orientation and polarization of the ''N''-acyl nucleophile. | The mutant Asp174Ala showed significantly decreased activity towards ''O''-aryl ''N''-acetylglucosaminides possessing either good or poor leaving groups and it was argued that this is consistent with its role as a residue responsible for the orientation and polarization of the ''N''-acyl nucleophile. | ||
| − | |||
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The reported crystallization of ''Clostridium perfringens'' NagJ (''Cp''OGA)<cite>ABB2005</cite> was followed by solved structures for that enzyme <cite>DvA2006</cite> and ''Bacteroides thetaiotaomicron'' ''β''-glucosaminidase (''Bt''OGA)<cite>DvA2006</cite>. In common with the chitinases of family GH18 and the ''exo''-acting ''β''-hexosaminidases of GH20 the catalytic domain is a (''βα'')<sub>8</sub>-barrel structure. The structure of human ''O''-GlcNAcase has not been solved but both bacterial homologues are good models of both the active site and catalytic domain of human ''O''-GlcNAcase. More recently the structure of the GH84 ''β''-hexosaminidase from ''Oceanicola granulosus'' has been solved and shown to possess good sequence identity with the catalytic domain of the human enzyme <cite>DvA2010</cite>. Furthermore, the ''C''-terminal domain of this protein also displays notable sequence identity with the spacer domain, which separates the domains possessing ''O''-GlcNAcase and histone acetyltransferase activities) of the human enzyme. | + | The reported crystallization of ''Clostridium perfringens'' NagJ (''Cp''OGA) <cite>ABB2005</cite> was followed by solved structures for that enzyme <cite>DvA2006</cite> and ''Bacteroides thetaiotaomicron'' ''β''-glucosaminidase (''Bt''OGA) <cite>DvA2006</cite>. In common with the chitinases of family [[GH18]] and the ''exo''-acting ''β''-hexosaminidases of [[GH20]] the catalytic domain is a (''βα'')<sub>8</sub>-barrel structure. The structure of human ''O''-GlcNAcase has not been solved but both bacterial homologues are good models of both the active site and catalytic domain of human ''O''-GlcNAcase. More recently the structure of the GH84 ''β''-hexosaminidase from ''Oceanicola granulosus'' has been solved and shown to possess good sequence identity with the catalytic domain of the human enzyme <cite>DvA2010</cite>. Furthermore, the ''C''-terminal domain of this protein also displays notable sequence identity with the spacer domain, which separates the domains possessing ''O''-GlcNAcase and histone acetyltransferase activities) of the human enzyme. |
[[Image:GH84conf.jpg|thumb|450px|'''Defining the conformational itinerary of a GH84 enzyme.''' Azepane (i) binds with a boat-like conformation to ''Bacteroides thetaiotaomicron'' ''β''-hexosaminidase (''Bt''OG). This binding mode is confirmed by the structure of a bound substrate (ii). Thiazoline (iii) binds to wild-type ''Bt''OG, 5-fluoro-oxazoline (iv) binds to the Asp243Asn mutant of ''Bt''OG, and oxazoline (v) binds to the Asp242Asn mutant of ''Bt''OG in the <sup>4</sup>C<sub>1</sub> conformation.]] | [[Image:GH84conf.jpg|thumb|450px|'''Defining the conformational itinerary of a GH84 enzyme.''' Azepane (i) binds with a boat-like conformation to ''Bacteroides thetaiotaomicron'' ''β''-hexosaminidase (''Bt''OG). This binding mode is confirmed by the structure of a bound substrate (ii). Thiazoline (iii) binds to wild-type ''Bt''OG, 5-fluoro-oxazoline (iv) binds to the Asp243Asn mutant of ''Bt''OG, and oxazoline (v) binds to the Asp242Asn mutant of ''Bt''OG in the <sup>4</sup>C<sub>1</sub> conformation.]] | ||
| − | A series of crystallographic studies on ''Bacteroides thetaiotaomicron'' ''β''-hexosaminidase have used a variety of small molecules to define the conformational itinerary for this family of enzymes. Substrate distortion from the stable <sup>4</sup>C<sub>1</sub> conformation found in solution to a bound <sup>1,4</sup>B / <sup>1</sup>S<sub>3</sub> conformation was supported by the crystal structure of the wild-type enzyme in complex with 6-acetamido-6-deoxy-castanospermine <cite>Mac2010</cite> and the 7-membered ring-containing azepane <cite>Ble2009</cite>. This distortion was confirmed by the structure the wild-type enzyme in complex with the substrate 3,4-difluorophenyl 2-deoxy-2-difluoroacetamido-''β''-D-glucopyranoside <cite>GJD2010</cite>. Early studies of the wild-type ''Bt''OGA-bound thiazoline show that this intermediate is found in a <sup>4</sup>C<sub>1</sub>conformation <cite>DJV2005</cite>; subsequent studies have shown that oxazoline intermediates are bound in this conformation to ''Bt''OGA active site mutants<cite>GJD2010</cite>. | + | A series of crystallographic studies on ''Bacteroides thetaiotaomicron'' ''β''-hexosaminidase have used a variety of small molecules to define the conformational itinerary for this family of enzymes. Substrate distortion from the stable <sup>4</sup>C<sub>1</sub> conformation found in solution to a bound <sup>1,4</sup>B / <sup>1</sup>S<sub>3</sub> conformation was supported by the crystal structure of the wild-type enzyme in complex with 6-acetamido-6-deoxy-castanospermine <cite>Mac2010</cite> and the 7-membered ring-containing azepane <cite>Ble2009</cite>. This distortion was confirmed by the structure the wild-type enzyme in complex with the substrate 3,4-difluorophenyl 2-deoxy-2-difluoroacetamido-''β''-D-glucopyranoside <cite>GJD2010</cite>. Early studies of the wild-type ''Bt''OGA-bound thiazoline show that this intermediate is found in a <sup>4</sup>C<sub>1</sub>conformation <cite>DJV2005</cite>; subsequent studies have shown that oxazoline intermediates are bound in this conformation to ''Bt''OGA active site mutants <cite>GJD2010</cite>. |
| − | |||
== Family Firsts == | == Family Firsts == | ||
| − | ;First sterochemistry determination: <sup>1</sup>H-NMR studies of human ''O''-GlcNAcase established that the ''β''-configured hemiacetal product | + | ;First sterochemistry determination: <sup>1</sup>H-NMR studies of human ''O''-GlcNAcase established that it hydrolyzes the product with [[retaining|retention]] of anomeric stereochemistry to give the ''β''-configured hemiacetal product, which then anomerises in solution <cite>DJV2009</cite>. |
| − | ;First catalytic nucleophile identification: This family of enzymes uses a mechanism of neighbouring group participation, | + | ;First [[catalytic nucleophile]] identification: This family of enzymes uses a mechanism of neighbouring group participation, in the the substrate ''N''-acetyl group acts as a nucleophile. This was first established through the use of free energy relationships studies <cite>DJV2005</cite>. |
| − | ;First general acid/base residue identification: Studies of human ''O''-GlcNAcase mutant Asp175Ala | + | ;First [[general acid/base]] residue identification: Studies of human ''O''-GlcNAcase mutant Asp175Ala identified reactivity patterns (free energy relationships, pH-activity profiles) consistent with the action of Asp175 as the catalytic [[general acid/base]] <cite>DJV2006</cite>. |
;First 3-D structure: The structures of ''Bacteroides thetaiotaomicron'' ''O''-GlcNAcase <cite>GJD2006</cite> and ''Clostridium perfringens'' NagJ <cite>DvA2006</cite>. | ;First 3-D structure: The structures of ''Bacteroides thetaiotaomicron'' ''O''-GlcNAcase <cite>GJD2006</cite> and ''Clostridium perfringens'' NagJ <cite>DvA2006</cite>. | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | |||
#Gao01 pmid=11148210 | #Gao01 pmid=11148210 | ||
#Guthrie89 Guthrie RD, Jencks WP. ''IUPAC Recommendations for the Representation of Reaction Mechanisms'' Acc. Chem. Res. 1989; 22(10): 343-349. | #Guthrie89 Guthrie RD, Jencks WP. ''IUPAC Recommendations for the Representation of Reaction Mechanisms'' Acc. Chem. Res. 1989; 22(10): 343-349. | ||
| Line 76: | Line 71: | ||
#DJV2009 pmid=19715310 | #DJV2009 pmid=19715310 | ||
#DJV2010 pmid=20067256 | #DJV2010 pmid=20067256 | ||
| − | #DJV2006 | + | #DJV2006 pmid=16533067 |
#GJD2006 pmid=16565725 | #GJD2006 pmid=16565725 | ||
#GJD2009 pmid=19217614 | #GJD2009 pmid=19217614 | ||
| Line 93: | Line 88: | ||
#Sato1999 pmid=9917327 | #Sato1999 pmid=9917327 | ||
#Mac2010 pmid=20851343 | #Mac2010 pmid=20851343 | ||
| − | |||
</biblio> | </biblio> | ||
[[Category:Glycoside Hydrolase Families|GH084]] | [[Category:Glycoside Hydrolase Families|GH084]] | ||
Latest revision as of 13:21, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH84 | |
| Clan | none |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH84.html | |
Substrate specificities
Glycoside hydrolases of GH84 include β-N-acetylglucosaminidases but members have often also been annotated as β-N-acetylhyaluronidases. This second annotation arises from the initial cloning of the enzyme and its early sequence analysis, which suggested sequence similarity to β-N-acetylhyaluronidases [1]. Biochemical analysis of a GH84 enzyme annotated in as a hyaluronidase, however, revealed that it had no such activity [2] and structural studies of other homologues have suggested the active site would be unable to process polymeric hyaluronan. Humans possess only one enzyme belonging to GH84, first defined by its unique biochemical properties including activity at neutral pH and selectivity for N-acetylglucosamine residues. The human GH84 enzyme was labelled as HexC to distinguish it from the human GH20 lysosomal enzymes HexA and HexB. HexC was first cloned from a meningoma library using autologous serum and defined as meningoma expressed antigen 5 (MGEA5) [1]. It was later cloned and biochemically characterized as O-GlcNAcase, a nuclear and cytoplasmic enzyme targeting glycoprotein substrates modified by β-linked GlcNAc residues linked to serine and threonine residues [3, 4]. A truncated, nuclear-localized isoform of human O-GlcNAcase lacking a putative C-terminal histone acetyl transferase domain retains similar kinetic properties (aside from a much lower activity) and inhibition patterns as the full-length cytosolic isoform and is consistent with hexosaminidase activity residing in the N-terminal domain, which shares similarity with other GH84 enzymes [5]. In contrast to the β-hexosaminidases of GH20 a relaxed specificity for substitutions of the N-acyl group is observed with residues significantly more bulky than the N-acyl group being tolerated [6].
Kinetics and Mechanism
Members of GH84 are retaining enzymes that use a catalytic mechanism of neighboring group participation, this originally being established through the use of free-energy relationship-based studies of human O-GlcNAcase [6]. More recent studies of the human enzyme have investigated variations in rates of reaction (V/K) with both nucleophile and leaving group structures [7]. For substrates possessing the naturally-occurring acetyl nucleophile a pre-chemical step is rate-determining on V/K for leaving groups with a pKa below 11 (with the chemical step rate-determining for substrates with higher pKa values). Studies of substrates possessing fluoroacetyl nucleophiles highlighted that a dissociative (DN*AN) [8] mechanism involving general acid catalysis operates for the hydrolysis of substrates possessing leaving groups with a pKa greater than approximately 7; a concerted (ANDN) [8] mechanism, not employing general acid catalysis was found for substrates possessing leaving groups with lower pKa values (consistent with prior studies showing hydrolysis of 1-S-glucosaminides [9]).
Numerous carbohydrate and carbohydrate like-scaffolds have been reported as potent inhibitors of GH84 enzymes. These include "NAG-thiazolines" [6], PUGNAc (O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino-N-phenylcarbamate) [10], GlcNAcstatins [11, 12], 6-epi-valeinamines [13], and 6-acetamido-6-deoxy-castanospermine [14]. The greater tolerance of the N-acyl binding pockets of GH84 enzymes for bulky sidechains compared to those of GH20 enzymes has enabled the development of inhibitors that are not only highly potent but also highly selective in their inhibition of human nucleocytoplasmic O-GlcNAcase (GH84) over human lysosomal β-hexosaminidases [6, 15, 16]. Many of these families of inhibitors possess structural characteristics reminiscent of the oxocarbenium ion-like transition states of glycosyl group transfer and, as such may loosely be termed 'transition state analogues'. An analysis of NAG-thiazoline- and PUGNAc-derived inhibitors of human O-GlcNAcase has shown that only the NAG-thiazolines position the inhibitors and their N-acyl side-chains within the hydrophobic binding pocket in a manner consistent with the species found along the reaction coordinate [15]. As such NAG-thiazoline inhibitors may be termed 'Bartlett-type' (free-energy relationship-based) [17] transition state analogues. Significantly streptozotocin, a widely used diabetogenic compound whose toxicity towards pancreatic β-cells was hypothesized to arise from its ability to act as an inhibitor of human O-GlcNAcase, neither binds covalently to GH84 enzymes [6, 18, 19], nor is a particularly potent inhibitor of human O-GlcNAcase [6, 20, 21].
Catalytic Residues
Studies of two mutants of human O-GlcNAcase established that adjacent aspartate residues, Asp174 and Asp175, act as critical components of the catalytic machinery of this enzyme [22].
The mutant Asp175Ala displayed marked reductions in activity (V and (V/K)) towards aryl N-acetylglucosaminides possessing poor leaving groups with smaller reductions being observed for both O-aryl and S-aryl N-acetylglucosaminides substrates possessing better leaving groups. Exogenous azide was found to partially rescue the activity of human O-GlcNAcase towards 3,4-dinitrophenylglucosaminide. These results clearly identified Asp175 as the general acid catalyst.
The mutant Asp174Ala showed significantly decreased activity towards O-aryl N-acetylglucosaminides possessing either good or poor leaving groups and it was argued that this is consistent with its role as a residue responsible for the orientation and polarization of the N-acyl nucleophile.
Three-dimensional structures
The reported crystallization of Clostridium perfringens NagJ (CpOGA) [23] was followed by solved structures for that enzyme [24] and Bacteroides thetaiotaomicron β-glucosaminidase (BtOGA) [24]. In common with the chitinases of family GH18 and the exo-acting β-hexosaminidases of GH20 the catalytic domain is a (βα)8-barrel structure. The structure of human O-GlcNAcase has not been solved but both bacterial homologues are good models of both the active site and catalytic domain of human O-GlcNAcase. More recently the structure of the GH84 β-hexosaminidase from Oceanicola granulosus has been solved and shown to possess good sequence identity with the catalytic domain of the human enzyme [25]. Furthermore, the C-terminal domain of this protein also displays notable sequence identity with the spacer domain, which separates the domains possessing O-GlcNAcase and histone acetyltransferase activities) of the human enzyme.
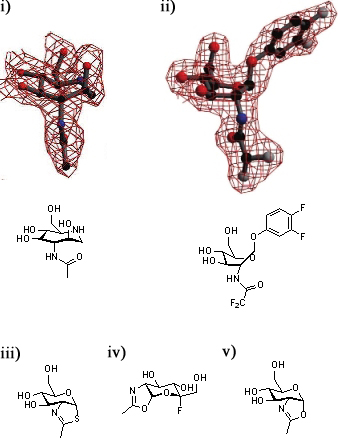
A series of crystallographic studies on Bacteroides thetaiotaomicron β-hexosaminidase have used a variety of small molecules to define the conformational itinerary for this family of enzymes. Substrate distortion from the stable 4C1 conformation found in solution to a bound 1,4B / 1S3 conformation was supported by the crystal structure of the wild-type enzyme in complex with 6-acetamido-6-deoxy-castanospermine [14] and the 7-membered ring-containing azepane [26]. This distortion was confirmed by the structure the wild-type enzyme in complex with the substrate 3,4-difluorophenyl 2-deoxy-2-difluoroacetamido-β-D-glucopyranoside [27]. Early studies of the wild-type BtOGA-bound thiazoline show that this intermediate is found in a 4C1conformation [6]; subsequent studies have shown that oxazoline intermediates are bound in this conformation to BtOGA active site mutants [27].
Family Firsts
- First sterochemistry determination
- 1H-NMR studies of human O-GlcNAcase established that it hydrolyzes the product with retention of anomeric stereochemistry to give the β-configured hemiacetal product, which then anomerises in solution [7].
- First catalytic nucleophile identification
- This family of enzymes uses a mechanism of neighbouring group participation, in the the substrate N-acetyl group acts as a nucleophile. This was first established through the use of free energy relationships studies [6].
- First general acid/base residue identification
- Studies of human O-GlcNAcase mutant Asp175Ala identified reactivity patterns (free energy relationships, pH-activity profiles) consistent with the action of Asp175 as the catalytic general acid/base [22].
- First 3-D structure
- The structures of Bacteroides thetaiotaomicron O-GlcNAcase [28] and Clostridium perfringens NagJ [24].
References
Error fetching PMID 16533067:
- Heckel D, Comtesse N, Brass N, Blin N, Zang KD, and Meese E. (1998). Novel immunogenic antigen homologous to hyaluronidase in meningioma. Hum Mol Genet. 1998;7(12):1859-72. DOI:10.1093/hmg/7.12.1859 |
- Sheldon WL, Macauley MS, Taylor EJ, Robinson CE, Charnock SJ, Davies GJ, Vocadlo DJ, and Black GW. (2006). Functional analysis of a group A streptococcal glycoside hydrolase Spy1600 from family 84 reveals it is a beta-N-acetylglucosaminidase and not a hyaluronidase. Biochem J. 2006;399(2):241-7. DOI:10.1042/BJ20060307 |
- Error fetching PMID 11148210:
- Dong DL and Hart GW. (1994). Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol Chem. 1994;269(30):19321-30. | Google Books | Open Library
- Macauley MS and Vocadlo DJ. (2009). Enzymatic characterization and inhibition of the nuclear variant of human O-GlcNAcase. Carbohydr Res. 2009;344(9):1079-84. DOI:10.1016/j.carres.2009.04.017 |
- Macauley MS, Whitworth GE, Debowski AW, Chin D, and Vocadlo DJ. (2005). O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280(27):25313-22. DOI:10.1074/jbc.M413819200 |
- Greig IR, Macauley MS, Williams IH, and Vocadlo DJ. (2009). Probing synergy between two catalytic strategies in the glycoside hydrolase O-GlcNAcase using multiple linear free energy relationships. J Am Chem Soc. 2009;131(37):13415-22. DOI:10.1021/ja904506u |
-
Guthrie RD, Jencks WP. IUPAC Recommendations for the Representation of Reaction Mechanisms Acc. Chem. Res. 1989; 22(10): 343-349.
- Macauley MS, Stubbs KA, and Vocadlo DJ. (2005). O-GlcNAcase catalyzes cleavage of thioglycosides without general acid catalysis. J Am Chem Soc. 2005;127(49):17202-3. DOI:10.1021/ja0567687 |
- Stubbs KA, Zhang N, and Vocadlo DJ. (2006). A divergent synthesis of 2-acyl derivatives of PUGNAc yields selective inhibitors of O-GlcNAcase. Org Biomol Chem. 2006;4(5):839-45. DOI:10.1039/b516273d |
- Dorfmueller HC, Borodkin VS, Schimpl M, Shepherd SM, Shpiro NA, and van Aalten DM. (2006). GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J Am Chem Soc. 2006;128(51):16484-5. DOI:10.1021/ja066743n |
- Shanmugasundaram B, Debowski AW, Dennis RJ, Davies GJ, Vocadlo DJ, and Vasella A. (2006). Inhibition of O-GlcNAcase by a gluco-configured nagstatin and a PUGNAc-imidazole hybrid inhibitor. Chem Commun (Camb). 2006(42):4372-4. DOI:10.1039/b612154c |
- Scaffidi A, Stubbs KA, Dennis RJ, Taylor EJ, Davies GJ, Vocadlo DJ, and Stick RV. (2007). A 1-acetamido derivative of 6-epi-valienamine: an inhibitor of a diverse group of beta-N-acetylglucosaminidases. Org Biomol Chem. 2007;5(18):3013-9. DOI:10.1039/b709681j |
- Macauley MS, He Y, Gloster TM, Stubbs KA, Davies GJ, and Vocadlo DJ. (2010). Inhibition of O-GlcNAcase using a potent and cell-permeable inhibitor does not induce insulin resistance in 3T3-L1 adipocytes. Chem Biol. 2010;17(9):937-48. DOI:10.1016/j.chembiol.2010.07.006 |
- Whitworth GE, Macauley MS, Stubbs KA, Dennis RJ, Taylor EJ, Davies GJ, Greig IR, and Vocadlo DJ. (2007). Analysis of PUGNAc and NAG-thiazoline as transition state analogues for human O-GlcNAcase: mechanistic and structural insights into inhibitor selectivity and transition state poise. J Am Chem Soc. 2007;129(3):635-44. DOI:10.1021/ja065697o |
- Dorfmueller HC, Borodkin VS, Schimpl M, Zheng X, Kime R, Read KD, and van Aalten DM. (2010). Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases. Chem Biol. 2010;17(11):1250-5. DOI:10.1016/j.chembiol.2010.09.014 |
- Mader MM and Bartlett PA. (1997). Binding Energy and Catalysis: The Implications for Transition-State Analogs and Catalytic Antibodies. Chem Rev. 1997;97(5):1281-1302. DOI:10.1021/cr960435y |
- Pathak S, Dorfmueller HC, Borodkin VS, and van Aalten DM. (2008). Chemical dissection of the link between streptozotocin, O-GlcNAc, and pancreatic cell death. Chem Biol. 2008;15(8):799-807. DOI:10.1016/j.chembiol.2008.06.010 |
- He Y, Martinez-Fleites C, Bubb A, Gloster TM, and Davies GJ. (2009). Structural insight into the mechanism of streptozotocin inhibition of O-GlcNAcase. Carbohydr Res. 2009;344(5):627-31. DOI:10.1016/j.carres.2008.12.007 |
- Konrad RJ, Mikolaenko I, Tolar JF, Liu K, and Kudlow JE. (2001). The potential mechanism of the diabetogenic action of streptozotocin: inhibition of pancreatic beta-cell O-GlcNAc-selective N-acetyl-beta-D-glucosaminidase. Biochem J. 2001;356(Pt 1):31-41. DOI:10.1042/0264-6021:3560031 |
- Hanover JA, Lai Z, Lee G, Lubas WA, and Sato SM. (1999). Elevated O-linked N-acetylglucosamine metabolism in pancreatic beta-cells. Arch Biochem Biophys. 1999;362(1):38-45. DOI:10.1006/abbi.1998.1016 |
- Error fetching PMID 16533067:
- Ficko-Blean E and Boraston AB. (2005). Cloning, recombinant production, crystallization and preliminary X-ray diffraction studies of a family 84 glycoside hydrolase from Clostridium perfringens. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61(Pt 9):834-6. DOI:10.1107/S1744309105024012 |
- Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, and van Aalten DM. (2006). Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25(7):1569-78. DOI:10.1038/sj.emboj.7601026 |
- Schimpl M, Schüttelkopf AW, Borodkin VS, and van Aalten DM. (2010). Human OGA binds substrates in a conserved peptide recognition groove. Biochem J. 2010;432(1):1-7. DOI:10.1042/BJ20101338 |
- Marcelo F, He Y, Yuzwa SA, Nieto L, Jiménez-Barbero J, Sollogoub M, Vocadlo DJ, Davies GD, and Blériot Y. (2009). Molecular basis for inhibition of GH84 glycoside hydrolases by substituted azepanes: conformational flexibility enables probing of substrate distortion. J Am Chem Soc. 2009;131(15):5390-2. DOI:10.1021/ja809776r |
- He Y, Macauley MS, Stubbs KA, Vocadlo DJ, and Davies GJ. (2010). Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J Am Chem Soc. 2010;132(6):1807-9. DOI:10.1021/ja9086769 |
- Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, and Davies GJ. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol. 2006;13(4):365-71. DOI:10.1038/nsmb1079 |
- He Y, Macauley MS, Stubbs KA, Vocadlo DJ, and Davies GJ. (2010). Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J Am Chem Soc. 2010;132(6):1807-9. DOI:10.1021/ja9086769 |