CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 99"
| (3 intermediate revisions by the same user not shown) | |||
| Line 19: | Line 19: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | [[File:Figure1.png|thumb|'''Figure 1. Carbohydrate array assay results of FvCBM99. ''' Binding analysis with porphyran and other galactans | + | [[File:Figure1.png|thumb|'''Figure 1. Carbohydrate array assay results of FvCBM99. ''' Binding analysis with porphyran and other galactans from red algae (A), and other polysaccharides (B). κ-Car, κ-carrageenan. ι-Car, ι-carrageenan. Alg, alginate. An-FUC, sulfated fucan from ''Ascophyllum nodosum''. CSA, chondroitin sulfate A sodium salt. HA, hyaluronic acid. Pec, pectin. Each polysaccharide performed three parallels. The values (noted at the bottom) were the average gray intensities of the triplicates. The value binding to porphyran was set to 100, and the other values were normalized accordingly.''' ]] |
| − | The first characterized member in the CBM99 family is FvCBM99 <cite>Mei2023</cite>. The CBM FvCBM99 | + | The first characterized member in the CBM99 family is FvCBM99 <cite>Mei2023</cite>. The CBM FvCBM99 bound to porphyran and displayed a weak affinity to agarose (Fig. 1). It was incapable of binding to the other examined polysaccharides, including κ-carrageenan, ι-carrageenan, alginate, sulfated fucan, chondroitin sulfate A sodium salt, hyaluronic acid, and pectin. Furthermore, FvCBM99 bound to porphyran tetrasaccharide with an affinity constant of 1.9 × 10<sup>-4</sup> M, but not to agarose tetrasaccharide. Since agarose chains usually contain a few characteristic structural units of porphyran <cite>Chi2012</cite>, it was thus speculated that the weak affinity of FvCBM99 to agarose was attributed to the structural heterogeneity of agarose. The polysaccharide and oligosaccharide binding assays show that FvCBM99 specifically binds to the major structural units of porphyran. |
== Structural Features == | == Structural Features == | ||
| − | + | An AlphaFold2 model predicts that FvCBM99 has a β-sandwich fold <cite>Mei2023</cite>. | |
== Functionalities == | == Functionalities == | ||
To evaluate the feasibility of FvCBM99 as a tool in the ''in situ'' investigation of porphyran, a fluorescent probe was constructed by fusing FvCBM99 with a green fluorescent protein. The ''in situ'' visualization of porphyran in red alga ''Porphyra haitanensis'' was realized by utilizing the fluorescent probe <cite>Mei2023</cite>. | To evaluate the feasibility of FvCBM99 as a tool in the ''in situ'' investigation of porphyran, a fluorescent probe was constructed by fusing FvCBM99 with a green fluorescent protein. The ''in situ'' visualization of porphyran in red alga ''Porphyra haitanensis'' was realized by utilizing the fluorescent probe <cite>Mei2023</cite>. | ||
| − | Most of the members of the CBM99 family are appended to the | + | |
| + | Most of the members of the CBM99 family are appended to the [[GH16]]_11, [[GH16]]_12, [[GH16]]_16, and [[GH16]]_26 subfamily, or the [[GH86]] family. As documented in the CAZy database, members of the four subfamilies and the [[GH86]] family exhibit enzymatic activity for degrading porphyran. It suggests that multiple porphyran-binding CBMs might be present in the CBM99 family. Furthermore, the homologs of FvCBM99, including WP_102755601.1 (located from 770 to 859 amino acids) and WP_108602097.1 (located from 720 to 822 amino acids), were shown to bind to porphyran. | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First Identified | + | ;First Identified: The first member FvCBM99 is a component of a potential [[GH86]] porphyranase (GenBank: AJW82063.1) from a marine bacterium ''Flammeovirga sp''. OC4 <cite>Liu2015</cite>. |
| − | The first member FvCBM99 is a component of a potential GH86 porphyranase (GenBank: AJW82063.1) from a marine bacterium ''Flammeovirga sp''. OC4 <cite>Liu2015</cite>. | + | ;First Structural Characterization: No experimental three-dimensional structure has been solved in this family. |
| − | ;First Structural Characterization | ||
| − | No three-dimensional structure has been solved in this | ||
== References == | == References == | ||
Latest revision as of 23:54, 15 February 2024
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM99.html |
Ligand specificities
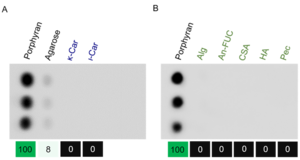
The first characterized member in the CBM99 family is FvCBM99 [1]. The CBM FvCBM99 bound to porphyran and displayed a weak affinity to agarose (Fig. 1). It was incapable of binding to the other examined polysaccharides, including κ-carrageenan, ι-carrageenan, alginate, sulfated fucan, chondroitin sulfate A sodium salt, hyaluronic acid, and pectin. Furthermore, FvCBM99 bound to porphyran tetrasaccharide with an affinity constant of 1.9 × 10-4 M, but not to agarose tetrasaccharide. Since agarose chains usually contain a few characteristic structural units of porphyran [2], it was thus speculated that the weak affinity of FvCBM99 to agarose was attributed to the structural heterogeneity of agarose. The polysaccharide and oligosaccharide binding assays show that FvCBM99 specifically binds to the major structural units of porphyran.
Structural Features
An AlphaFold2 model predicts that FvCBM99 has a β-sandwich fold [1].
Functionalities
To evaluate the feasibility of FvCBM99 as a tool in the in situ investigation of porphyran, a fluorescent probe was constructed by fusing FvCBM99 with a green fluorescent protein. The in situ visualization of porphyran in red alga Porphyra haitanensis was realized by utilizing the fluorescent probe [1].
Most of the members of the CBM99 family are appended to the GH16_11, GH16_12, GH16_16, and GH16_26 subfamily, or the GH86 family. As documented in the CAZy database, members of the four subfamilies and the GH86 family exhibit enzymatic activity for degrading porphyran. It suggests that multiple porphyran-binding CBMs might be present in the CBM99 family. Furthermore, the homologs of FvCBM99, including WP_102755601.1 (located from 770 to 859 amino acids) and WP_108602097.1 (located from 720 to 822 amino acids), were shown to bind to porphyran.
Family Firsts
- First Identified
- The first member FvCBM99 is a component of a potential GH86 porphyranase (GenBank: AJW82063.1) from a marine bacterium Flammeovirga sp. OC4 [3].
- First Structural Characterization
- No experimental three-dimensional structure has been solved in this family.
References
- Mei X, Zhang Y, Liu G, Shen J, Han J, Xue C, Xiao H, and Chang Y. (2023). Characterization of a novel carbohydrate-binding module specifically binding to the major structural units of porphyran. Int J Biol Macromol. 2023;253(Pt 5):127106. DOI:10.1016/j.ijbiomac.2023.127106 |
- Chi WJ, Chang YK, and Hong SK. (2012). Agar degradation by microorganisms and agar-degrading enzymes. Appl Microbiol Biotechnol. 2012;94(4):917-30. DOI:10.1007/s00253-012-4023-2 |
- Liu Y, Yi Z, Cai Y, and Zeng R. (2015). Draft genome sequence of algal polysaccharides degradation bacterium, Flammeovirga sp. OC4. Mar Genomics. 2015;21:21-2. DOI:10.1016/j.margen.2015.02.001 |