CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycosyltransferase Family 47"
| Line 56: | Line 56: | ||
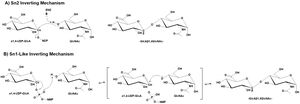
GT47 enzymes employ an inverting catalytic mechanism where the hydroxyl group of an acceptor substrate presumably acts as a nucleophile in a SN2 single displacement reaction. The result is an inversion of the anomeric configuration of the transferred sugar from an α-linked sugar nucleotide donor to form a β-linked extended glycan product. While an SN2 mechanism would predict the deprotonation of the acceptor nucleophile by an enzyme associated catalytic base, the structure of the EXT1 active site did not appear to contain an appropriately positioned ionizable group to act as catalytic base <cite>LiH2023</cite>. Similar structural studies on the inverting GT-B fold glycosyltransferases, POFUT1 <cite>LiZ2017 Lira2018 Lira2011</cite> and AtFUT1 <cite>Urbanowicz2017</cite>, also indicated the lack of an appropriately positioned catalytic base for deprotonation. In these latter cases a non-canonical SN1-like mechanism was proposed. A similar SN1-like mechanism may also occur for the GT47 enzymes <cite>Zhang2023 Moremen2019</cite>. | GT47 enzymes employ an inverting catalytic mechanism where the hydroxyl group of an acceptor substrate presumably acts as a nucleophile in a SN2 single displacement reaction. The result is an inversion of the anomeric configuration of the transferred sugar from an α-linked sugar nucleotide donor to form a β-linked extended glycan product. While an SN2 mechanism would predict the deprotonation of the acceptor nucleophile by an enzyme associated catalytic base, the structure of the EXT1 active site did not appear to contain an appropriately positioned ionizable group to act as catalytic base <cite>LiH2023</cite>. Similar structural studies on the inverting GT-B fold glycosyltransferases, POFUT1 <cite>LiZ2017 Lira2018 Lira2011</cite> and AtFUT1 <cite>Urbanowicz2017</cite>, also indicated the lack of an appropriately positioned catalytic base for deprotonation. In these latter cases a non-canonical SN1-like mechanism was proposed. A similar SN1-like mechanism may also occur for the GT47 enzymes <cite>Zhang2023 Moremen2019</cite>. | ||
| − | [[File: | + | [[File:GT47_mechanism_V7.jpg|thumb|300px|right|'''Figure 1: Proposed Mechanism of GT47 Domain in EXT1.''' GT47 enzymes employ an inverting catalytic mechanism through two potential inverting mechanisms. A) In the SN2 inverting mechanism, the hydroxyl group of an acceptor substrate (shown as GlcNAc) acts as nucleophile in a single displacement reaction leading to inversion in anomeric configuration of the transferred sugar. B) The lack of an appropriately positioned ionizable group to act as catalytic base in the EXT1 structure suggested a non-canonical SN1-like mechanism for the GT47 domain of EXT1. A similar SN1-like mechanism may also occur for other GT47 enzymes.]] |
== Catalytic Residues == | == Catalytic Residues == | ||
Revision as of 12:51, 12 July 2024
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: Daniel Tehrani and Charlie Corulli
- Responsible Curators: Breeanna Urbanowicz and Kelley Moremen
| Glycosyltransferase Family GT47 | |
| Fold | GT-B |
| Mechanism | Inverting |
| Active site residues | Known/unknown |
| CAZy DB link | |
| https://www.cazy.org/GT47.html | |
Substrate specificities
Glycosyltransferases in GT47 catalyze the transfer of a wide variety of monosaccharides from activated donor sugar nucleotides onto a diversity of acceptor substrates found in plants, animals, insects, and bacteria [1]. Donor sugar nucleotides for discrete clades of GT47 enzymes include UDP-Arabinofuranose (UDP-Araf), UDP-Arabinopyranose (UDP-Arap), UDP-Xylose (UDP-Xyl), UDP-Galactose (UDP-Gal), UDP-Galacturonic acid (UDP-GalA), and UDP-Glucuronic acid (UDP-GlcA, i.e. EXT1) [1, 2, 3, 4, 5, 6, 7]. Genes encoding members of the GT47 family are found across all domains of life, and known biochemical pathways GT47 enzymes include diverse plant cell wall polysaccharides and glycoproteins and the heparan sulfate backbone [1, 7].
Plants
The GT47 family is highly diversified in plants, having an association with the biosynthesis of almost every class of plant cell wall polysaccharide [1]. Although the enzymatic functions for the vast majority of plant GT47s are currently unknown, analysis of plant mutants has identified members predicted to use UDP-GalA, UDP-Gal, UDP-Arap, UDP-Araf, and UDP-Xyl as activated sugar donors. Known acceptor polysaccharide substrates include xyloglucan, xylan, galacto-glucomannan, xylo-galacturonan, cell wall extensins, and rhamnogalacturonan I. Most members of GT47 from plants have been identified through analysis of mutants.
Xyloglucan
Xyloglucan is a hemicellulose and a major component of primary cell walls of dicots [8]. Xyloglucan forms polymer-polymer interactions with cellulose that can be influenced by the diversity of sidechains found on xyloglucan that vary depending on plant species, tissue, and stage of growth [9]. Various members of the GT47 family have been reported to contribute to the synthesis of the numerous sidechains found on xyloglucan, with the two most notable being the xyloglucan-modifying galactosyltransferases MURUS3 (MUR3) and Xyloglucan L-Side Chain Galactosyltransferase2 (XLT2). These enzymes catalyze the regiospecific addition of β-D-Gal forming the Gal-β1,2-Xyl-α- (‘L) sidechains of xyloglucan [5, 10]. XyG “S”-Side Chain Transferase1 (XST1) and Xyloglucan “D”‐Side Chain Transferase (XDT)are reported to transfer UDP-Araf and UDP-Arap respectively to the 3rd, reducing end, xylose of xyloglucan, forming the Araf-α1,2-Xyl-α- (‘S) and Arap-α1,2- Xyl-α- (‘D) sidechain motifs [11, 12]. Xyloglucan-Specific Galacturonosyltransferase1 (XUT1) is reported to transfer UDP-GalA, forming the GalA-β1,2-Xyl-α- (‘Y) sidechain [6]. More recently, Xyloglucan Beta-Xylopyranosyltransferase (XBT) has been identified to transfer UDP-Xyl to form the Xyl-β1,2-Xyl-α-(‘U) sidechain [13]. Taken together, the quantity of GT47s identified to act on xyloglucan to date is indicative of the important role this family has in contributing to the diversity of this polymer.
Xylan
Unlike the previously mentioned sidechain modifications of xyloglucan, GT47s can additionally contribute to the synthesis of polymer backbones as observed with xylan. Xylan is a hemicellulosic polysaccharide and a major component of plant secondary cell walls. This polysaccharide is composed of a β1,4-Xyl backbone that is directly synthesized by Xylan Synthase (XYS) enzymes. They are thought to function in a complex with two other members of GT43 [1, 14, 15]. This xylan synthase complex (XSC) contributes to the synthesis of the xylan backbone, although XYS is the only enzyme in the complex which displays an enzymatic function in extending xylan in vivo. Loss of function mutations have additionally identified Irregular Xylem7 (IRX7) as another potential xylan modifying GT47, hypothesized to participate in synthesis of the reducing end tetrasaccharide β-D-Xyl-1,4-[β-D-Xyl-1,3-α-l-Rha-1,2-α-D-GalA-1,4-D-Xyl] present in xylans from dicots, although more evidence is required to elucidate this function [14, 15].
Mannan
Mannan is a hemicellulosic polysaccharide prominently found in the plant primary cell wall. Galactoglucomannan is a classification of mannan with a backbone interspersed with β1,4-Glc which can be further substituted with α1,6-Gal residues. Recently, it was shown that the α1,6-Gal residues of this polymer can additionally be substituted with β1,2-Gal. Loss of function mutations in Arabidopsis have identified the mannan β-galactosyltransferase (MBGT) as the most likely candidate in synthesizing the Galβ-1,2-Galα-1,6- sidechains, catalyzing addition of the terminal galactose to the structure [16].
Pectin
Pectin encompasses a diverse group of polymers which include homogalacturonan, rhamnogalacturonan I, rhamnogalacturonan II, and xylogalacturonan. Pectic polysaccharides play many crucial roles in plants such as intercellular adhesion, stress response, seed germination, morphogenesis, and cell communication [1, 17]. Loss of function mutations in Arabidopsis have identified Xylogalacturonan Deficient1 (XGD1) as a xylosyltransferase catalyzing the addition of β1,4-Xyl residues onto homogalacturonan backbone to form xylogalacturonan [18]. Arabinan Deficient 1 (ARAD1) likely contributes to the synthesis of arabinan sidechains of rhamnogalacturonan I, and was identified via analysis of isolated RG-I from arad1 Arabidopsis mutants [3].
Extensin
Unlike the previously mentioned polysaccharides, extensins are rod-like hydroxyproline rich glycoproteins (HRGP) that form crosslinked networks in the plant cell wall. These networks are reported to play a crucial role in regulating cell wall growth and development [19]. A unique member of the GT47 family, Extensin Arabinose Deficient transferase (ExAD), is reported to synthesize the addition of the fourth arabinofuranose (Araf) on Araf substituted C4-hydroxyprolines (Hyps) creating Hyp-Araf4, a unique feature found on extensins [19, 20].
Animals
The abundance of GT47 family enzymes in mammals is more restricted and includes only members of the Exostosin (EXT) and Exostoslin-Like (EXTL) family of enzymes involved in heparan sulfate biosynthesis. Heparan sulfate is comprised of a repeat disaccharide polymer of ( GlcAβ1,4GlcNAcα1,4-)n that is further elaborated with extensive sulfation along the polymer chain. The disaccharide backbone repeat is elongated by the co-polymerase activity of the heterodimeric EXT1-EXT2 complex [7]. EXT1 and EXT2 are homologous two domain enzymes, and each protein chain contains a GT47 β1,4-GlcA transferase-like and a GT64 α1,4GlcNAc transferase-like domain. Surprisingly, only the GT47 domain of EXT1 and GT64 domain of EXT2 exhibit catalytic activity, while the other domains in each subunit are nonfunctional [7]. Additional EXT homologs include the EXTL proteins, EXTL1-3. EXTL1 and EXTL3 are two domain proteins, each harboring a GT47 and GT64 domain like EXT1 and EXT2. However, only the GT64 domains exhibit α1,4GlcNAc transferase activity, while their corresponding GT47 domains are inactive. In contrast, EXTL2 is a single GT64 domain enzyme with a α1,4GlcNAc transferase activity, while the corresponding GT47 domain present in other EXTs is missing. Thus, among the five mammalian EXT or EXTL homologs, only EXT1 contains a functional GT47 domain exhibiting β1,4-GlcA transferase activity.
Kinetics and Mechanism
GT47 enzymes employ an inverting catalytic mechanism where the hydroxyl group of an acceptor substrate presumably acts as a nucleophile in a SN2 single displacement reaction. The result is an inversion of the anomeric configuration of the transferred sugar from an α-linked sugar nucleotide donor to form a β-linked extended glycan product. While an SN2 mechanism would predict the deprotonation of the acceptor nucleophile by an enzyme associated catalytic base, the structure of the EXT1 active site did not appear to contain an appropriately positioned ionizable group to act as catalytic base [7]. Similar structural studies on the inverting GT-B fold glycosyltransferases, POFUT1 [21, 22, 23] and AtFUT1 [24], also indicated the lack of an appropriately positioned catalytic base for deprotonation. In these latter cases a non-canonical SN1-like mechanism was proposed. A similar SN1-like mechanism may also occur for the GT47 enzymes [1, 25].
Catalytic Residues
Unlike GT-A fold enzymes, GT-B fold enzymes like GT47s lack the predictable catalytic features, such as a DxD motif, G-loop, xED, and C-term His, that are involved in sugar nucleotide and divalent cation interactions [26]. In place of the bridging interactions between the nucleotide sugar donor diphosphate residues and an enzyme bound divalent cation as found in GT-A fold enzymes, GT-B fold glycosyltransferases employ basic Lys and Arg side chains for interaction with the diphosphate [1, 25, 27]. Mutation of the active site Lys and Arg residues in the GT47 domain of EXT1 completely eliminated β1,4-GlcA transferase activity as well as co-polymerase activity for extension of heparan sulfate backbone synthesis [7]. Additional residues involved in donor and acceptor interactions were identified in the EXT1:UDP:acceptor complex during structural studies, but further mutagenesis studies were not performed to test function [7]. Analogous Lys and Arg residues can be identified in the putative donor binding sites in AlphaFold models plant GT47 enzymes, but their roles in catalysis have not been tested.
Three-dimensional structures
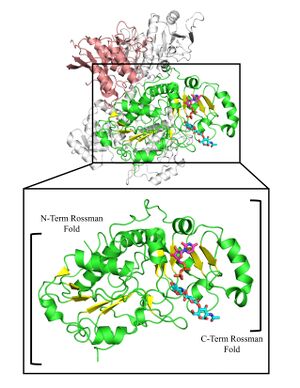
GT47 enzymes are characterized by GT-B fold architecture comprised of two linked Rossmann-fold domains with a cleft between the domains containing the active site. GT47 enzymes bind their nucleotide sugar donor through interactions with the C-terminal Rossmann fold domain, while the acceptor substrate generally binds either in the cleft between the two domains or exclusively with the N-terminal Rossmann fold domain. The binding sites for donor and acceptor residues are generally comprised of loop regions extending from the respective Rossmann fold domains facing toward the cleft between the two domains [1, 25, 27].
Family Firsts
The first structure of a CAZy family GT47 was the cryo-EM structure of the human EXT1-2 heterocomplex containing a GT47 β1,4-GlcA transferase domain and an inactive GT64 α1,4GlcNAc transferase-like domain of EXT1, while EXT2 contains an inactive GT47 β1,4-GlcA transferase-like domain along with an active GT64 α1,4GlcNAc transferase domain [7]. Structures of UDP and acceptor co-complexes were determined for each of the enzyme active sites to map substrate interactions. The structures provided insight into the overall enzyme fold (GT-B) and catalytic site structure and mechanism (inverting) as a framework for studies on the other CAZy GT47 enzymes, especially the GT47s in plants that lack empirical structures.
References
-
Zhang L, Prabhakar Pradeep K, Bharadwaj Vivek S, Bomble Yannick J, Peña Maria J, Urbanowicz Breeanna R. (2023) Glycosyltransferase family 47 (GT47) proteins in plants and animals. Essays in Biochemistry. 2023;67(3):639-52.DOI:10.1042/EBC20220152.
- Li X, Cordero I, Caplan J, Mølhøj M, and Reiter WD. (2004). Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol. 2004;134(3):940-50. DOI:10.1104/pp.103.036285 |
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, and Scheller HV. (2006). ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 2006;140(1):49-58. DOI:10.1104/pp.105.072744 |
- Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, and Marchant A. (2009). The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 2009;57(4):718-31. DOI:10.1111/j.1365-313X.2008.03724.x |
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, and Reiter WD. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell. 2003;15(7):1662-70. DOI:10.1105/tpc.009837 |
- Peña MJ, Kong Y, York WS, and O'Neill MA. (2012). A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell. 2012;24(11):4511-24. DOI:10.1105/tpc.112.103390 |
- Li H, Chapla D, Amos RA, Ramiah A, Moremen KW, and Li H. (2023). Structural basis for heparan sulfate co-polymerase action by the EXT1-2 complex. Nat Chem Biol. 2023;19(5):565-574. DOI:10.1038/s41589-022-01220-2 |
- Zabotina OA (2012). Xyloglucan and its biosynthesis. Front Plant Sci. 2012;3:134. DOI:10.3389/fpls.2012.00134 |
- Schultink A, Liu L, Zhu L, and Pauly M. (2014). Structural Diversity and Function of Xyloglucan Sidechain Substituents. Plants (Basel). 2014;3(4):526-42. DOI:10.3390/plants3040526 |
-
Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M. (2012) RNA-Seq Analysis of Developing Nasturtium Seeds (Tropaeolum majus): Identification and Characterization of an Additional Galactosyltransferase Involved in Xyloglucan Biosynthesis. Molecular Plant. 2012;5(5):984-92.DOI:10.1093/mp/sss032.
-
Schultink A, Cheng K, Park YB, Cosgrove DJ, Pauly M. (2013) The Identification of Two Arabinosyltransferases from Tomato Reveals Functional Equivalency of Xyloglucan Side Chain Substituents. Plant Physiology. 2013;163(1):86-94.DOI: 10.1104/pp.113.221788
- Zhu L, Dama M, and Pauly M. (2018). Identification of an arabinopyranosyltransferase from Physcomitrella patens involved in the synthesis of the hemicellulose xyloglucan. Plant Direct. 2018;2(3):e00046. DOI:10.1002/pld3.46 |
- Immelmann R, Gawenda N, Ramírez V, and Pauly M. (2023). Identification of a xyloglucan beta-xylopyranosyltransferase from Vaccinium corymbosum. Plant Direct. 2023;7(7):e514. DOI:10.1002/pld3.514 |
- Brown DM, Zhang Z, Stephens E, Dupree P, and Turner SR. (2009). Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009;57(4):732-46. DOI:10.1111/j.1365-313X.2008.03729.x |
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, and Turner SR. (2007). Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007;52(6):1154-68. DOI:10.1111/j.1365-313X.2007.03307.x |
- Yu L, Yoshimi Y, Cresswell R, Wightman R, Lyczakowski JJ, Wilson LFL, Ishida K, Stott K, Yu X, Charalambous S, Wurman-Rodrich J, Terrett OM, Brown SP, Dupree R, Temple H, Krogh KBRM, and Dupree P. (2022). Eudicot primary cell wall glucomannan is related in synthesis, structure, and function to xyloglucan. Plant Cell. 2022;34(11):4600-4622. DOI:10.1093/plcell/koac238 |
- Shin Y, Chane A, Jung M, and Lee Y. (2021). Recent Advances in Understanding the Roles of Pectin as an Active Participant in Plant Signaling Networks. Plants (Basel). 2021;10(8). DOI:10.3390/plants10081712 |
- Jensen JK, Sørensen SO, Harholt J, Geshi N, Sakuragi Y, Møller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, Pauly M, Beldman G, Willats WG, and Scheller HV. (2008). Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell. 2008;20(5):1289-302. DOI:10.1105/tpc.107.050906 |
- Showalter AM and Basu D. (2016). Extensin and Arabinogalactan-Protein Biosynthesis: Glycosyltransferases, Research Challenges, and Biosensors. Front Plant Sci. 2016;7:814. DOI:10.3389/fpls.2016.00814 |
- Møller SR, Yi X, Velásquez SM, Gille S, Hansen PLM, Poulsen CP, Olsen CE, Rejzek M, Parsons H, Yang Z, Wandall HH, Clausen H, Field RA, Pauly M, Estevez JM, Harholt J, Ulvskov P, and Petersen BL. (2017). Identification and evolution of a plant cell wall specific glycoprotein glycosyl transferase, ExAD. Sci Rep. 2017;7:45341. DOI:10.1038/srep45341 |
- Li Z, Han K, Pak JE, Satkunarajah M, Zhou D, and Rini JM. (2017). Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat Chem Biol. 2017;13(7):757-763. DOI:10.1038/nchembio.2381 |
- Lira-Navarrete E and Hurtado-Guerrero R. (2018). A perspective on structural and mechanistic aspects of protein O-fucosylation. Acta Crystallogr F Struct Biol Commun. 2018;74(Pt 8):443-450. DOI:10.1107/S2053230X18004788 |
- Lira-Navarrete E, Valero-González J, Villanueva R, Martínez-Júlvez M, Tejero T, Merino P, Panjikar S, and Hurtado-Guerrero R. (2011). Structural insights into the mechanism of protein O-fucosylation. PLoS One. 2011;6(9):e25365. DOI:10.1371/journal.pone.0025365 |
- Urbanowicz BR, Bharadwaj VS, Alahuhta M, Peña MJ, Lunin VV, Bomble YJ, Wang S, Yang JY, Tuomivaara ST, Himmel ME, Moremen KW, York WS, and Crowley MF. (2017). Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism. Plant J. 2017;91(6):931-949. DOI:10.1111/tpj.13628 |
- Moremen KW and Haltiwanger RS. (2019). Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat Chem Biol. 2019;15(9):853-864. DOI:10.1038/s41589-019-0350-2 |
-
Taujale R, Venkat A, Huang L-C, Zhou Z, Yeung W, Rasheed KM, Li S, Edison AS, Moremen KW, Kannan N. (2020) Deep evolutionary analysis reveals the design principles of fold A glycosyltransferases. eLife. 2020;9:e54532.DOI:10.7554/eLife.54532.
-
Rini JM, Moremen KW, Davis BG, Esko JD. (2022) Glycosyltransferases and Glycan-Processing Enzymes. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH, editors. Essentials of Glycobiology. 4th ed. Cold Spring Harbor (NY)2022. p. 67-78.DOI 10.1101/glycobiology.4e.6.