CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 20"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) |
||
| Line 41: | Line 41: | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
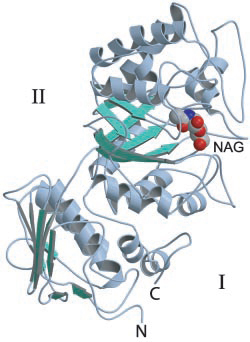
| − | The first GH20 enzyme to have its structure determined was the ''Serratia marscescens'' chitobiase | + | The first GH20 enzyme to have its structure determined was the ''Serratia marscescens'' chitobiase <cite>Tews1996</cite>. This enzyme's active site is located at the C-terminal end of the third of four protein domains, a (''βα'')<sub>8</sub>-barrel. On the basis of a structure of this enzyme in complex with the substrate chitobiose, the invariant Glu540 was identified as the likely catalytic general acid/base. Furthermore the ''N''-acetyl group of the non-reducing ''N''-acetylglucosamine residue was found to have its carbonyl oxygen atom suitably positioned to act as the nucleophile.<cite>SJW2002</cite> [[Image:Sphex.jpg|thumb|300px|'''Ribbon diagram of ''SpHex'' with "NAG-thiazoline" bound in active site.''' The catalytic domain II is a (''βα'')<sub>8</sub>-barrel.]] |
| − | |||
== Family Firsts == | == Family Firsts == | ||
Revision as of 23:53, 24 February 2011
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Ian Greig^^^
- Responsible Curator: ^^^David Vocadlo^^^
| Glycoside Hydrolase Family GH20 | |
| Clan | GH-K |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/fam/GH20.html | |
Substrate specificities
GH20 members comprise enzymes from both eukaryotes and prokaryotes. In addition to exo-acting β-N-acetylglucosaminidases, β-N-acetylgalactosamindase and β-6-SO3-N-acetylglucosaminidases, GH20 also contains exo-acting lacto-N-biosidases that cleave β-D-Gal-(1→3)-D-GlcNAc disaccharides from the non-reducing end of oligosaccharides. The best known members of this family are the human isoenzymes hexosaminidase A (a heterodimer of α and β subunits) and B (a homodimer of β subunits), which are responsible for the hydrolysis of the terminal GalNAc residue from the GM2 ganglioside (GalNAcβ(1–4)-[NANAα(2–3)-]-Galβ(1–4)-Glc-ceramide) within the lysosome. Mutations to these enzymes are responsible for the lysosomal storage disorders Tay-Sachs disease (HEXA) and Sandhoff disease (HEXB). Inhibitors of these enzymes are being developed as chemical chaperones to promote the partial restoration of enzyme activity in vivo and treat these genetic disorders [1].
Kinetics and Mechanism
Neighbouring group participation has long been established as a reasonable mechanism for glycoside hydrolysis in solution[2, 3, 4, 5] and originally outlined as a possible, though subsequently refuted, mechanism for the hen egg-white lysozyme-catalyzed cleavage of β-aryl di-N-acetylchitobiosides [6]. The earliest kinetic evidence supporting a mechanism involving neighbouring group participation in an enzyme-catalyzed hydrolysis [7, 8] can be found for an N-acetyl-β-D-glucosaminidase isolated from Aspergillus oryzae [9], likely a GH20 enzyme. This work used free energy relationships to infer neighbouring group participation although complete Michaelis-Menten kinetic parameters were not determined. Such kinetic parameters were determined for a β-N-acetylglucosaminidase from Aspergillus niger and a similar free energy relationship-based analysis carried out to infer neighbouring group participation for this enzyme which, though unknown, is likely from GH20 [10]. The potency of "NAG-thiazoline" as a competitive inhibitor of the jack bean N-acetyl-β-D-hexosaminidase (Ki = 280 nM) has also been used to infer a mechanism of neighbouring group participation although, interestingly, the only retaining hexosaminidases reported currently (November 2010) reported in the CAZy database for the genus Canavalia are found in GH18 [11].
Deacetylation of the non-reducing end of a series of chito-oligosaccharides results in a loss of activity of Serratia marscescens chitobiase, an established GH20 enzyme, towards these compounds, which instead act as competitive inhibitors.[12] Moreover the structure of Serratia marcescens chitobiase in complex with a substrate provides structural support for a substrate-assisted mechanism.
A comparative analysis of the activity of Streptomyces plicatus β-hexosaminidase (SpHex, GH20) and Vibrio furnisii β-hexosaminidase (ExoII, GH3) towards p-nitrophenyl N-acyl glucosaminides highlights contrasting reactivity trends expected for families of β-glucosaminidase utilizing a mechanism of substrate-assisted catalysis (GH20) and those which do not (GH3): sharp decreases in activity with increasing N-acyl fluorination are observed in the case of the SpHex enzyme whereas negligible changes in activity are observed for ExoII [13].
Catalytic Residues
The key catalytic residues of GH20 enzymes are found in conserved D-E amino acid pair. This catalytic diad is preceded in the primary sequence by the consensus H-x-G-G motif. The glutamate residue functions as the catalytic general acid/base. As these enzymes employ neighbouring group participation the preceding aspartate is not a nucleophile. Rather kinetic and crystallographic studies have shown that this residue orients and polarizes the catalytic N-acetyl residue [14]. It may function either as a general base by deprotonating the N-acetyl group in the intermediate and forming a neutral oxazoline intermediate, or alternatively it may electrostatically stabilize a positively charge oxazolinium ion intermediate. The catalytic N-acetyl group of the substrate is bound in a hydrophobic pocket defined by three conserved tryptophan residues. These three tryptophan residues define a compact pocket which does not accommodate (non-native) extended N-acyl side-chains as readily as the elongated hydrophobic pocket found in GH84 enzymes [15].
Three-dimensional structures
The first GH20 enzyme to have its structure determined was the Serratia marscescens chitobiase [16]. This enzyme's active site is located at the C-terminal end of the third of four protein domains, a (βα)8-barrel. On the basis of a structure of this enzyme in complex with the substrate chitobiose, the invariant Glu540 was identified as the likely catalytic general acid/base. Furthermore the N-acetyl group of the non-reducing N-acetylglucosamine residue was found to have its carbonyl oxygen atom suitably positioned to act as the nucleophile.[14]
Family Firsts
- First sterochemistry determination
- The first stereochemical determination for a known member of GH20 was on the Serratia marscescens enzyme[12]. The stereochemistry of hydrolysis of three different hexosaminidases (human placenta, jack bean, and bovine kidney) was shown by the Withers group in 1994 [17] and it is now generally assumed that some of these are GH20 enzymes.
- First catalytic nucleophile identification
- These enzymes employ neighbouring group participation. Prior to the advent of the CAZy system of classification, kinetic studies of the (likely GH20) β-N-hexosaminidases from Aspergillus oryzae[9] and Aspergillus niger[10] supported such a mechanism. This mechanism was further suggested by both the 3-D structure of Serratia marcescens chitobiase [16] (by analogy with GH18 enzymes), through work in which the non-reducing end sugar was de-acetylated resulting in total loss in activity [12], and by potent inhibition of Jack Bean β-hexosaminidase by NAG-thiazoline[11].
- First general acid/base residue identification
- Inferred from the 3-D structure [16] and by analogy with structurally related GH18 chitinases.
- First 3-D structure
- The 3-D structure of the Serratia marscescens chitobiase [16].
References
- Tropak MB, Reid SP, Guiral M, Withers SG, and Mahuran D. (2004). Pharmacological enhancement of beta-hexosaminidase activity in fibroblasts from adult Tay-Sachs and Sandhoff Patients. J Biol Chem. 2004;279(14):13478-87. DOI:10.1074/jbc.M308523200 |
-
Cocker, D, Sinnott, ML (1976) Acetolysis of 2,4-Dinitrophenyl Glycopyranosides. J. C. S. Perkin II 90, 618-620.
-
Piszkiewicz, D, Bruice, T (1967) Glycoside Hydrolysis. I. Intramolecular Acetamido and Hydroxyl Group Catalysis in Glycoside Hydrolysis. J. Am. Chem. Soc. 89, 6237-6243.
-
Piszkiewicz, D, Bruice, T (1968) Glycoside Hydrolysis. II. Intramolecular Carboxyl and Acetamido Group Catalysis in β-Glycoside Hydrolysis. J. Am. Chem. Soc. 90, 2156-2163.
-
Piszkiewicz, D, Bruice, T (1968) Glycoside Hydrolysis. III. Intramolecular Acetamido Group Participation in the Specific Acid Catalyzed Hydrolysis of Methyl-2-Acetamido-2-deoxy-β-D-glucopyranoside. J. Am. Chem. Soc. 90, 5844-5848.
-
Lowe, G, Sheppard, G, Sinnott, ML, Williams, A, (1967) Lysozyme-Catalysed Hydrolysis of some 'β-Aryl Di-N-acetylchitobiosides. Biochem J. 104(3), 893-899.
-
Yamamoto, K, (1973) N-Acyl Specificity of Taka-N-acetyl-β-D-glucosaminidase Studied by Synthetic Substrate Analogs II. Preparation of Some p-Nitrophenyl 2-Halogenoacetylamino-2-deoxy-β-D-glucopyranoside and Their Susceptibility to Enzymic Hydrolysis. J. Biochem. 73, 749-753.
-
Yamamoto, K, (1974) A Quantitative Approach to the Evaluation of β-Acetamide Substituent Effects on the Hydrolysis by Taka-N-acetyl-β-D-glucosaminidase. Role of the Substrate 2-Acetamide Group in the N-Acyl Specificity of the Enzyme J. Biochem. 76, 385-390.
-
Mega, T, Ikenaka, T, Matsushima, Y, (1970) Studies on N-Acetyl-β-D-glucosaminidase of Aspergillus oryzae. J. Biochem. 68, 109-117.
-
Jones, CS, Kosman, DJ (1980) Purification, Properties, Kinetics, and Mechanism of β-N-Acetylglucosaminidase from Aspergillus niger. J. Biol. Chem. 255(24), 11861-11869.
-
Knapp, S, Vocadlo, DJ, Gao, Z, Kirk, B, Lou, J, Withers, SG (1996) NAG-thiazoline, An N-Acetyl-β-hexosaminidase Inhibitor That Implicates Acetamido Participation. J. Am. Chem. Soc. 118, 6804-6805.
- Drouillard S, Armand S, Davies GJ, Vorgias CE, and Henrissat B. (1997). Serratia marcescens chitobiase is a retaining glycosidase utilizing substrate acetamido group participation. Biochem J. 1997;328 ( Pt 3)(Pt 3):945-9. DOI:10.1042/bj3280945 |
- Vocadlo DJ and Withers SG. (2005). Detailed comparative analysis of the catalytic mechanisms of beta-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry. 2005;44(38):12809-18. DOI:10.1021/bi051121k |
- Williams SJ, Mark BL, Vocadlo DJ, James MN, and Withers SG. (2002). Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J Biol Chem. 2002;277(42):40055-65. DOI:10.1074/jbc.M206481200 |
- Macauley MS, Whitworth GE, Debowski AW, Chin D, and Vocadlo DJ. (2005). O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280(27):25313-22. DOI:10.1074/jbc.M413819200 |
- Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson KS, and Vorgias CE. (1996). Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol. 1996;3(7):638-48. DOI:10.1038/nsb0796-638 |
- Lai EC and Withers SG. (1994). Stereochemistry and kinetics of the hydration of 2-acetamido-D-glucal by beta-N-acetylhexosaminidases. Biochemistry. 1994;33(49):14743-9. DOI:10.1021/bi00253a012 |
- Comfort DA, Bobrov KS, Ivanen DR, Shabalin KA, Harris JM, Kulminskaya AA, Brumer H, and Kelly RM. (2007). Biochemical analysis of Thermotoga maritima GH36 alpha-galactosidase (TmGalA) confirms the mechanistic commonality of clan GH-D glycoside hydrolases. Biochemistry. 2007;46(11):3319-30. DOI:10.1021/bi061521n |
- He S and Withers SG. (1997). Assignment of sweet almond beta-glucosidase as a family 1 glycosidase and identification of its active site nucleophile. J Biol Chem. 1997;272(40):24864-7. DOI:10.1074/jbc.272.40.24864 |
- Robert V. Stick and Spencer J. Williams. (2009) Carbohydrates. Elsevier Science.
-
Sinnott, M.L. (1990) Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev. 90, 1171-1202. DOI: 10.1021/cr00105a006