CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 3"
(Created page with "<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> {{UnderConstruct...") |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (23 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Harry Gilbert|Harry Gilbert]] and [[User:Ed Bayer|Ed Bayer]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Ed Bayer|Ed Bayer]] |
---- | ---- | ||
| Line 11: | Line 11: | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
|- | |- | ||
| − | | colspan="2" |{{CAZyDBlink}} | + | | colspan="2" |{{CAZyDBlink}}CBM3.html |
|} | |} | ||
</div> | </div> | ||
<!-- This is the end of the table --> | <!-- This is the end of the table --> | ||
| − | |||
== Ligand specificities == | == Ligand specificities == | ||
| − | + | CBM3 is a Gram-positive bacterial family of protein modules that comprise around 150 amino acids. The family is divided into four subgroups, CBM3a-d. The major ligand recognised by CBM3as and CBM3bs is crystalline cellulose with an affinity (K<sub>D</sub>) of 0.4 µM determined by depletion isotherms <cite>Morag1995</cite>. Isothermal titration calorimetry showed that binding to crystalline cellulose was entropically driven consistent with apolar interactions resulting in the release of caged water molecules from a ligand with a restricted conformation <cite>Hernandez-Gomez2015</cite>. CBM3s that bind to crystalline cellulose also interact with chitin and xyloglucan with an affinity ~500 lower than for crystalline cellulose. | |
| − | '' | + | Interaction with soluble xyloglucans by CBM3s was enthalpically driven with changes in entropy having a negative impact on affinity <cite>Hernandez-Gomez2015</cite>. The site of binding of a CBM3 from the ''Clostridium thermocellulum'' scaffoldin (CipA) to crystalline cellulose was determined by transmission electron microscopy with detection of the protein by immuno-gold labelling. The data showed that the CBM3 bound to the 110 face of Valonia cellulose <cite>Lehtio2003</cite>. The binding profile and site of cellulose recognition show that CBM3s are [[Carbohydrate-binding_modules#Types|type A]] modules. The three CBM3s from anti-sigma sensors displayed different specificities; Cthe_0059 CBM3b bound to a range of plant cell wall polysaccharides (PCWPs), Cthe_0404 CBM3b interacted weakly to xyloglucan but not to any other PCWP, and Cthe_0267 CBM3 bound primarily to crystalline and amorphous cellulose <cite>Nataf2010,Yaniv2014</cite>. |
== Structural Features == | == Structural Features == | ||
| − | '' | + | |
| − | + | [[File:cbm3SURFACE12.png|thumb|300px|right|'''Figure 1.''' The fold of the cellulose binding CBM3a from the ''Clostridium thermocellum'' scaffoldin CipA ([{{PDBlink}}1NBC PDB ID 1NBC]), highlighting the planar ligand binding site comprising five residues. The structure is rotated 90 degrees to illustrate the location of these residues on the β-sheet.]] | |
| − | + | ||
| − | + | The crystal structure of CBM3 from the C. thermocellum scaffoldin CipA revealed a classical β-jelly-roll fold consisting of nine β-strands in two antiparallel β-sheets comprising four (1, 2, 7, 4; β-sheet 1) and five (9, 8, 3, 6, 5; β-sheet 2) β strands, respectively <cite>Tormo1996</cite> ([{{PDBlink}}1NBC PDB ID 1NBC]). β-sheet 1 forms a flat surface that contains a linear array of five residues that presents a planar hydrophobic surface comprising a His, Trp, Tyr and an Arg-Asp ion pair ('''Figure 1'''). The residues in the planar strip were predicted to make apolar interactions with glucose molecules n, n+1, n+3 and n+5, consistent with mutagenesis data showing that each of the five amino acids played an important role in binding cellulose <cite>Benhar2001</cite>. Structures of CBMbs from other cellulosome-producing species followed that reinforced the original structural findings <cite>Shimon2000,Yaniv2011,Yaniv2012b</cite>. | |
| + | |||
| + | In other CBM3 modules that bind to cellulose, such as in the anti-σ-cell surface sensor RsgI1 (Cthe_0059), the His and ion pair are replaced by a Tyr and Phe, thus the hydrophobic planar binding site comprises four aromatic amino acids <cite>Yaniv2014</cite>. In a second cellulose-binding CBM3 located in an Rsgl sensor (Rsgl2, Cthe_0267), the aromatic planar strip is truncated, but lies planar with a hydrophobic protruding loop that is predicted to contribute to the cellulose binding site of the protein, similar to a group d CBM3 present in a GH48 exo-cellulase <cite>Cai2011</cite>. In addition to the hydrophobic strips it has also been proposed that highly conserved polar residues may be able to make productive hydrogen bonds with two additional cellulose chains in the microfibril <cite>Tormo1996,Yaniv2014</cite>. | ||
| + | |||
| + | In contrast to the planar face presented by β-sheet 1, β-sheet 2 displays a concave surface or shallow groove that contains highly conserved aromatic residues <cite>Tormo1996</cite>, suggesting that these hydrophobic amino acids are functionally significant. It has been proposed that the shallow cleft is involved in binding Pro-Thr linker segments and thus may contribute to structural organization of these multimodular proteins <cite>Yaniv2012a</cite>. | ||
== Functionalities == | == Functionalities == | ||
| − | + | CBM3s are derived from the scaffoldins <cite>Poole1992</cite> (non-catalytic proteins that that play an integral role in the assembly of multienzyme plant cell wall degrading complexes termed cellulosomes (see <cite>Fontes2010</cite> for review), sensor proteins that detect cellulose <cite>Nataf2010</cite> and a range of cellulases (e.g. <cite>Brunecky2013,Gilad2003,Sakon1997</cite>). In general CBM3s are separated from the other modules in these proteins by Pro-Thr-rich linker sequences. In some instances, however, group c CBM3 members (CBM3cs) are integral components of the substrate binding cleft of GH9 cellulases (e.g. <cite>Jindou2006,Burstein2009,Gilad2003,Sakon1997</cite>) In these enzymes the CBM3c modules, as discrete entities, do not bind to cellulose (reflecting the lack of conserved ligand binding residues) but play a pivotal role in the capacity of the cellulases to attack crystalline forms of the polysaccharide <cite>Sakon1997</cite>. It was proposed that the replacement of aromatic residues with conserved polar amino acids, altered the function of CBM3cs from an anchoring role. In the model proposed the polar residues in CBM3s replace the inter-chain hydrogen bonds within crystalline cellulose. The resultant disruption of the crystalline polysaccharide releases the cellulose chain on the centre of the CBM3c, which could then be fed into the active site cleft of the catalytic domain. Several studies have shown that CBM3 modules have enhanced the activity of cellulases <cite>Fierobe2002</cite> and a range of other plant cell wall degrading enzymes <cite>Zhang2014,Herve2010</cite>. These modules have also been used to probe the structure of plant cell walls <cite>Blake2006,Ding2006</cite>. | |
| − | |||
| − | |||
| − | |||
== Family Firsts == | == Family Firsts == | ||
| − | ;First Identified | + | ;First Identified: The first CBM3 to be identified (CipA-CBM3) was from the ''C. thermocellum'' scaffoldin CipA <cite>Poole1992</cite>. |
| − | : | + | |
| − | ;First Structural Characterization | + | ;First Structural Characterization: The first crystal structure of a CBM3, indeed of any CBM, is CipA-CBM3 <cite>Tormo1996</cite>. |
| − | : | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Morag1995 pmid=7646033 |
| − | # | + | #Hernandez-Gomez2015 pmid=26193423 |
| − | # | + | #Lehtio2003 pmid=12522267 |
| − | # | + | #Tormo1996 pmid=8918451 |
| − | # | + | #Benhar2001 Benhar, I., Tamarkin, A., Marash, L., Berdichevsky, Y., Yaron, S., Shoham, Y., Lamed, R., and Bayer, E. A. (2001) Phage display of cellulose binding domains for biotechnological application. In Glycosyl Hydrolases for Biomass Conversion (Himmel, M. E., Baker, J. O., and Saddler, J. N., Eds.), pp 168-189, American Chemical Society, Washington, DC. [http://dx.doi.org/10.1021/bk-2001-0769.ch010 DOI:10.1021/bk-2001-0769.ch010]. |
| − | # | + | #Shimon2000 pmid=11092922 |
| + | #Yaniv2011 pmid=21636890 | ||
| + | #Yaniv2012b pmid=22232162 | ||
| + | #Yaniv2014 pmid=24531486 | ||
| + | #Cai2011 pmid=21803997 | ||
| + | |||
| + | #Poole1992 pmid=1490597 | ||
| + | |||
| + | #Fontes2010 pmid=20373916 | ||
| + | #Nataf2010 pmid=20937888 | ||
| + | #Brunecky2013 pmid=24357319 | ||
| + | #Gilad2003 pmid=12511483 | ||
| + | #Sakon1997 pmid=9334746 | ||
| + | |||
| + | #Yaniv2012a pmid=22608730 | ||
| + | |||
| + | #Jindou2006 pmid=16445761 | ||
| + | #Burstein2009 pmid=19302786 | ||
| + | #Fierobe2002 pmid=12397074 | ||
| + | #Zhang2014 pmid=24297170 | ||
| + | #Herve2010 pmid=20696902 | ||
| + | #Blake2006 pmid=16844685 | ||
| + | #Ding2006 pmid=17068959 | ||
</biblio> | </biblio> | ||
[[Category:Carbohydrate Binding Module Families|CBM003]] | [[Category:Carbohydrate Binding Module Families|CBM003]] | ||
Latest revision as of 13:20, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM3.html |
Ligand specificities
CBM3 is a Gram-positive bacterial family of protein modules that comprise around 150 amino acids. The family is divided into four subgroups, CBM3a-d. The major ligand recognised by CBM3as and CBM3bs is crystalline cellulose with an affinity (KD) of 0.4 µM determined by depletion isotherms [1]. Isothermal titration calorimetry showed that binding to crystalline cellulose was entropically driven consistent with apolar interactions resulting in the release of caged water molecules from a ligand with a restricted conformation [2]. CBM3s that bind to crystalline cellulose also interact with chitin and xyloglucan with an affinity ~500 lower than for crystalline cellulose.
Interaction with soluble xyloglucans by CBM3s was enthalpically driven with changes in entropy having a negative impact on affinity [2]. The site of binding of a CBM3 from the Clostridium thermocellulum scaffoldin (CipA) to crystalline cellulose was determined by transmission electron microscopy with detection of the protein by immuno-gold labelling. The data showed that the CBM3 bound to the 110 face of Valonia cellulose [3]. The binding profile and site of cellulose recognition show that CBM3s are type A modules. The three CBM3s from anti-sigma sensors displayed different specificities; Cthe_0059 CBM3b bound to a range of plant cell wall polysaccharides (PCWPs), Cthe_0404 CBM3b interacted weakly to xyloglucan but not to any other PCWP, and Cthe_0267 CBM3 bound primarily to crystalline and amorphous cellulose [4, 5].
Structural Features
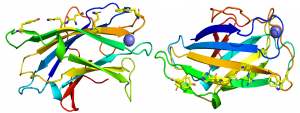
The crystal structure of CBM3 from the C. thermocellum scaffoldin CipA revealed a classical β-jelly-roll fold consisting of nine β-strands in two antiparallel β-sheets comprising four (1, 2, 7, 4; β-sheet 1) and five (9, 8, 3, 6, 5; β-sheet 2) β strands, respectively [6] (PDB ID 1NBC). β-sheet 1 forms a flat surface that contains a linear array of five residues that presents a planar hydrophobic surface comprising a His, Trp, Tyr and an Arg-Asp ion pair (Figure 1). The residues in the planar strip were predicted to make apolar interactions with glucose molecules n, n+1, n+3 and n+5, consistent with mutagenesis data showing that each of the five amino acids played an important role in binding cellulose [7]. Structures of CBMbs from other cellulosome-producing species followed that reinforced the original structural findings [8, 9, 10].
In other CBM3 modules that bind to cellulose, such as in the anti-σ-cell surface sensor RsgI1 (Cthe_0059), the His and ion pair are replaced by a Tyr and Phe, thus the hydrophobic planar binding site comprises four aromatic amino acids [5]. In a second cellulose-binding CBM3 located in an Rsgl sensor (Rsgl2, Cthe_0267), the aromatic planar strip is truncated, but lies planar with a hydrophobic protruding loop that is predicted to contribute to the cellulose binding site of the protein, similar to a group d CBM3 present in a GH48 exo-cellulase [11]. In addition to the hydrophobic strips it has also been proposed that highly conserved polar residues may be able to make productive hydrogen bonds with two additional cellulose chains in the microfibril [5, 6].
In contrast to the planar face presented by β-sheet 1, β-sheet 2 displays a concave surface or shallow groove that contains highly conserved aromatic residues [6], suggesting that these hydrophobic amino acids are functionally significant. It has been proposed that the shallow cleft is involved in binding Pro-Thr linker segments and thus may contribute to structural organization of these multimodular proteins [12].
Functionalities
CBM3s are derived from the scaffoldins [13] (non-catalytic proteins that that play an integral role in the assembly of multienzyme plant cell wall degrading complexes termed cellulosomes (see [14] for review), sensor proteins that detect cellulose [4] and a range of cellulases (e.g. [15, 16, 17]). In general CBM3s are separated from the other modules in these proteins by Pro-Thr-rich linker sequences. In some instances, however, group c CBM3 members (CBM3cs) are integral components of the substrate binding cleft of GH9 cellulases (e.g. [16, 17, 18, 19]) In these enzymes the CBM3c modules, as discrete entities, do not bind to cellulose (reflecting the lack of conserved ligand binding residues) but play a pivotal role in the capacity of the cellulases to attack crystalline forms of the polysaccharide [17]. It was proposed that the replacement of aromatic residues with conserved polar amino acids, altered the function of CBM3cs from an anchoring role. In the model proposed the polar residues in CBM3s replace the inter-chain hydrogen bonds within crystalline cellulose. The resultant disruption of the crystalline polysaccharide releases the cellulose chain on the centre of the CBM3c, which could then be fed into the active site cleft of the catalytic domain. Several studies have shown that CBM3 modules have enhanced the activity of cellulases [20] and a range of other plant cell wall degrading enzymes [21, 22]. These modules have also been used to probe the structure of plant cell walls [23, 24].
Family Firsts
- First Identified
- The first CBM3 to be identified (CipA-CBM3) was from the C. thermocellum scaffoldin CipA [13].
- First Structural Characterization
- The first crystal structure of a CBM3, indeed of any CBM, is CipA-CBM3 [6].
References
- Morag E, Lapidot A, Govorko D, Lamed R, Wilchek M, Bayer EA, and Shoham Y. (1995). Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl Environ Microbiol. 1995;61(5):1980-6. DOI:10.1128/aem.61.5.1980-1986.1995 |
- Hernandez-Gomez MC, Rydahl MG, Rogowski A, Morland C, Cartmell A, Crouch L, Labourel A, Fontes CM, Willats WG, Gilbert HJ, and Knox JP. (2015). Recognition of xyloglucan by the crystalline cellulose-binding site of a family 3a carbohydrate-binding module. FEBS Lett. 2015;589(18):2297-303. DOI:10.1016/j.febslet.2015.07.009 |
- Lehtiö J, Sugiyama J, Gustavsson M, Fransson L, Linder M, and Teeri TT. (2003). The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc Natl Acad Sci U S A. 2003;100(2):484-9. DOI:10.1073/pnas.212651999 |
- Nataf Y, Bahari L, Kahel-Raifer H, Borovok I, Lamed R, Bayer EA, Sonenshein AL, and Shoham Y. (2010). Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc Natl Acad Sci U S A. 2010;107(43):18646-51. DOI:10.1073/pnas.1012175107 |
- Yaniv O, Fichman G, Borovok I, Shoham Y, Bayer EA, Lamed R, Shimon LJ, and Frolow F. (2014). Fine-structural variance of family 3 carbohydrate-binding modules as extracellular biomass-sensing components of Clostridium thermocellum anti-σI factors. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 2):522-34. DOI:10.1107/S139900471302926X |
- Tormo J, Lamed R, Chirino AJ, Morag E, Bayer EA, Shoham Y, and Steitz TA. (1996). Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15(21):5739-51. | Google Books | Open Library
-
Benhar, I., Tamarkin, A., Marash, L., Berdichevsky, Y., Yaron, S., Shoham, Y., Lamed, R., and Bayer, E. A. (2001) Phage display of cellulose binding domains for biotechnological application. In Glycosyl Hydrolases for Biomass Conversion (Himmel, M. E., Baker, J. O., and Saddler, J. N., Eds.), pp 168-189, American Chemical Society, Washington, DC. DOI:10.1021/bk-2001-0769.ch010.
- Shimon LJ, Pagès S, Belaich A, Belaich JP, Bayer EA, Lamed R, Shoham Y, and Frolow F. (2000). Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 12):1560-8. DOI:10.1107/s0907444900012889 |
- Yaniv O, Shimon LJ, Bayer EA, Lamed R, and Frolow F. (2011). Scaffoldin-borne family 3b carbohydrate-binding module from the cellulosome of Bacteroides cellulosolvens: structural diversity and significance of calcium for carbohydrate binding. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 6):506-15. DOI:10.1107/S0907444911011322 |
- Yaniv O, Halfon Y, Shimon LJ, Bayer EA, Lamed R, and Frolow F. (2012). Structure of CBM3b of the major cellulosomal scaffoldin subunit ScaA from Acetivibrio cellulolyticus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 1):8-13. DOI:10.1107/S174430911104807X |
- Cai S, Zheng X, and Dong X. (2011). CBM3d, a novel subfamily of family 3 carbohydrate-binding modules identified in Cel48A exoglucanase of Cellulosilyticum ruminicola. J Bacteriol. 2011;193(19):5199-206. DOI:10.1128/JB.05227-11 |
- Yaniv O, Frolow F, Levy-Assraf M, Lamed R, and Bayer EA. (2012). Interactions between family 3 carbohydrate binding modules (CBMs) and cellulosomal linker peptides. Methods Enzymol. 2012;510:247-59. DOI:10.1016/B978-0-12-415931-0.00013-6 |
- Poole DM, Morag E, Lamed R, Bayer EA, Hazlewood GP, and Gilbert HJ. (1992). Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol Lett. 1992;78(2-3):181-6. DOI:10.1016/0378-1097(92)90022-g |
- Fontes CM and Gilbert HJ. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem. 2010;79:655-81. DOI:10.1146/annurev-biochem-091208-085603 |
- Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, Yang SJ, Resch MG, Adams MW, Lunin VV, Himmel ME, and Bomble YJ. (2013). Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science. 2013;342(6165):1513-6. DOI:10.1126/science.1244273 |
- Gilad R, Rabinovich L, Yaron S, Bayer EA, Lamed R, Gilbert HJ, and Shoham Y. (2003). CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J Bacteriol. 2003;185(2):391-8. DOI:10.1128/JB.185.2.391-398.2003 |
- Sakon J, Irwin D, Wilson DB, and Karplus PA. (1997). Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat Struct Biol. 1997;4(10):810-8. DOI:10.1038/nsb1097-810 |
- Jindou S, Xu Q, Kenig R, Shulman M, Shoham Y, Bayer EA, and Lamed R. (2006). Novel architecture of family-9 glycoside hydrolases identified in cellulosomal enzymes of Acetivibrio cellulolyticus and Clostridium thermocellum. FEMS Microbiol Lett. 2006;254(2):308-16. DOI:10.1111/j.1574-6968.2005.00040.x |
- Burstein T, Shulman M, Jindou S, Petkun S, Frolow F, Shoham Y, Bayer EA, and Lamed R. (2009). Physical association of the catalytic and helper modules of a family-9 glycoside hydrolase is essential for activity. FEBS Lett. 2009;583(5):879-84. DOI:10.1016/j.febslet.2009.02.013 |
- Fierobe HP, Bayer EA, Tardif C, Czjzek M, Mechaly A, Bélaïch A, Lamed R, Shoham Y, and Bélaïch JP. (2002). Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277(51):49621-30. DOI:10.1074/jbc.M207672200 |
- Zhang X, Rogowski A, Zhao L, Hahn MG, Avci U, Knox JP, and Gilbert HJ. (2014). Understanding how the complex molecular architecture of mannan-degrading hydrolases contributes to plant cell wall degradation. J Biol Chem. 2014;289(4):2002-12. DOI:10.1074/jbc.M113.527770 |
- Hervé C, Rogowski A, Blake AW, Marcus SE, Gilbert HJ, and Knox JP. (2010). Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A. 2010;107(34):15293-8. DOI:10.1073/pnas.1005732107 |
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, and Knox JP. (2006). Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem. 2006;281(39):29321-9. DOI:10.1074/jbc.M605903200 |
- Ding SY, Xu Q, Ali MK, Baker JO, Bayer EA, Barak Y, Lamed R, Sugiyama J, Rumbles G, and Himmel ME. (2006). Versatile derivatives of carbohydrate-binding modules for imaging of complex carbohydrates approaching the molecular level of resolution. Biotechniques. 2006;41(4):435-6, 438, 440 passim. DOI:10.2144/000112244 |