CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycosyltransferase Family 38"
Harry Brumer (talk | contribs) (Fixed broken link to Sorting the Diverse - new DOI link added) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (24 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| + | |||
{{UnderConstruction}} | {{UnderConstruction}} | ||
| − | * [[Author]]: | + | |
| − | * [[Responsible Curator]]: | + | * [[Author]]: [[User:Warren Wakarchuk|Warren Wakarchuk]] |
| + | * [[Responsible Curator]]: [[User:Warren Wakarchuk|Warren Wakarchuk]] | ||
| + | |||
---- | ---- | ||
<!-- The data in the table below should be updated by the Author/Curator according to current information on the family --> | <!-- The data in the table below should be updated by the Author/Curator according to current information on the family --> | ||
| − | <div style="float:right"> | + | <div style="float: right"> |
| − | {| {{Prettytable}} | + | {| {{Prettytable}} |
|- | |- | ||
| − | |{{Hl2}} colspan="2" align="center" |'''Glycosyltransferase Family GT38''' | + | | {{Hl2}} colspan="2" align="center" |'''Glycosyltransferase Family GT38''' |
|- | |- | ||
| − | |'''Clan''' | + | | '''Clan''' |
| − | | | + | | GT-B |
|- | |- | ||
| − | |''' | + | | '''Mechanisn''' |
| − | | | + | | inverting |
|- | |- | ||
| − | |'''Active site residues''' | + | | '''Active site residues''' |
| − | | | + | | known |
|- | |- | ||
| − | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | + | | {{Hl2}} colspan="2" align="center" |'''CAZy DB link''' |
|- | |- | ||
| colspan="2" |{{CAZyDBlink}}GT38.html | | colspan="2" |{{CAZyDBlink}}GT38.html | ||
|} | |} | ||
</div> | </div> | ||
| + | |||
<!-- This is the end of the table --> | <!-- This is the end of the table --> | ||
| − | |||
== Substrate specificities == | == Substrate specificities == | ||
| − | |||
| − | + | Members of GT-38 are the bacterial polysialyltransferases (polySTs), which catalyze the addition of sialic acids from the activated sugar donor, CMP-sialic acid (CMP-Neu5Ac), to the nonreducing end of the growing polySia chain <cite>Cho1994</cite>. These enzymes build the polymer as a capsular polysaccharide on a specialized poly-β-KDO modified lyso-phosphatidyl glycerol anchor in the membrane of Gram negative bacteria <cite>Willis2013</cite>. Bacterial polySia capsules exist in three different flavours: ''Escherichia coli'' K1, ''Neisseria meningitidis'' serotype B, ''Moraxella nonliquefaciens'', and ''Mannheimia haemolytica'' A2 synthesize α-2,8-linked polySia whereas ''N. meningitidis'' serotype C produces a α-2,9-linked polymer and ''E. coli'' K92 produces polymers with alternating α-2,8 and α-2,9 linkages <cite>Jennings1977 PuentePolledo Devi1991 Glode1977</cite>. ''In vitro'' enzyme reactions have shown that the members of GT-38 require two sialic acids for elongation <cite>Willis2008 Peterson2011 Lindhout2013</cite>, presumably as this mimics the ''in vivo'' lipid primer. The enzymes have been used in applications to modify therapeutic proteins and prepare synthetic vaccines <cite>Lindhout2011 McCarthy2013</cite>, where un-natural acceptors like protein N-glycans have been used. | |
| − | |||
| − | In the | ||
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | + | Sialic acid transfer occurs with inversion of configuration (from the β-linked CMP-Neu5Ac donor to the α-2,8-linked polySia), and polyST has been proposed to follow a SN2-like direct displacement mechanism. While H291 could act as a catalytic acid to stabilize the nucleotide phosphate-leaving group. | |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | + | Residues involved in catalysis have been proposed from site-directed mutagensis and the X-ray crystal structure from the ''M. hemolytica'' serotype A2 enzyme <cite>Lizak2017</cite>. The catalytic base E153 abstracts a proton from the C8′ hydroxyl group of the sialic acid acceptor concerted with the nucleophilic attack on the anomeric C2′ carbon of the CMP-sialic acid donor substrate, thereby generating an α-2,8 glycosidic linkage. The resulting negatively charged CMP leaving group is stabilized by H291 assisted by S339 and T340. | |
== Three-dimensional structures == | == Three-dimensional structures == | ||
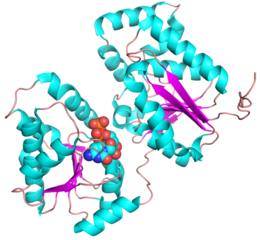
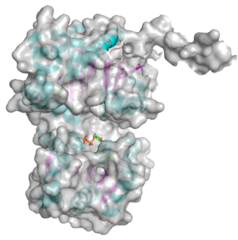
| − | + | ||
| + | <gallery widths=320px heights=240px perrow=2 caption="PDB ID 5WC6 from GT38 (click images for large versions)"> | ||
| + | File:GT38 3.png | ||
| + | File:Surface 2 cropped.png | ||
| + | |||
| + | </gallery> | ||
| + | |||
| + | The structure is a GT-B fold typical of a metal independent glycosyltransferase, it has 2 non-equivalent Rossmann-like folds. The structure was solved from a truncated version of the enzyme, which lacks 20 amino acids from the N-terminal end. There are two protomers in the crystal structure, but biochemical evidence suggests the soluble enzyme exists as a monomer. There is a hinge region between these domains (F227 to N236) which gives some flexibility in the structure. The structure shows an N-terminal tail which is unstructured and is likely to be a linker to the membrane anchor. The structure has a large electropositive groove which accomodates the polySia chain (right hand panel). One of the additional structures obtained for this enzyme is a complex with the synthetic heparin fondaparinux which was a surrogate for the polyanionic polySia. The image shown here is the CDP donor analogue complex which sites on one the Rossmann like domains. | ||
== Family Firsts == | == Family Firsts == | ||
| − | + | ||
| − | + | ; First general acid/base residue identification: E153, H291 | |
| − | ;First general acid/base residue identification: | + | |
| − | ;First 3-D structure: | + | Prior to having the structure, it had been suggested that the motif "HP" was involved in catalysis. Site directed mutagenesis with the MhpolyST suggested that the H291 residue does play an essential role in catalysis - as the general acid. This "HP" motif is also conserved in other bacterial sialytransferases. Similarly, the E153 residue has been proposed as the general base for this reaction. Both E153 and H291 show large decreases in kcat/Km when mutated to alanine - consistent with that assessment. |
| + | |||
| + | ; First 3-D structure: The family of structures are: PDB codes 5WC8, 5WCN, 5WC6 and 5WD7. | ||
| + | There are a family of structures, an apo-structure 5WC8, an acceptor complex (Sia2LacNAc6S) 5WCN, a donor analogue (CDP)5WC6, and a product analogue complex (fondapariux) 5WD7. | ||
| + | |||
| + | All of these structures required a mutation to decrease surface entropy - K69A. | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | # Cho1994 pmid=7972078 |
| − | # | + | # Willis2013 pmid=23610430 |
| + | # Jennings1977 pmid=408435 | ||
| + | # PuentePolledo pmid=10052589 | ||
| + | # Devi1991 pmid=1898915 | ||
| + | # Glode1977 pmid=64575 | ||
| + | # Lindhout2013 pmid=23922842 | ||
| + | # Peterson2011 pmid=21278299 | ||
| + | # Lindhout2011 pmid=21502532 | ||
| + | # McCarthy2013 pmid=23949787 | ||
| + | # Willis2008 pmid=18000029 | ||
| + | # Lizak2017 pmid=28724897 | ||
</biblio> | </biblio> | ||
| − | |||
[[Category:Glycosyltransferase Families|GT038]] | [[Category:Glycosyltransferase Families|GT038]] | ||
Latest revision as of 13:18, 18 December 2021
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| Glycosyltransferase Family GT38 | |
| Clan | GT-B |
| Mechanisn | inverting |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GT38.html | |
Substrate specificities
Members of GT-38 are the bacterial polysialyltransferases (polySTs), which catalyze the addition of sialic acids from the activated sugar donor, CMP-sialic acid (CMP-Neu5Ac), to the nonreducing end of the growing polySia chain [1]. These enzymes build the polymer as a capsular polysaccharide on a specialized poly-β-KDO modified lyso-phosphatidyl glycerol anchor in the membrane of Gram negative bacteria [2]. Bacterial polySia capsules exist in three different flavours: Escherichia coli K1, Neisseria meningitidis serotype B, Moraxella nonliquefaciens, and Mannheimia haemolytica A2 synthesize α-2,8-linked polySia whereas N. meningitidis serotype C produces a α-2,9-linked polymer and E. coli K92 produces polymers with alternating α-2,8 and α-2,9 linkages [3, 4, 5, 6]. In vitro enzyme reactions have shown that the members of GT-38 require two sialic acids for elongation [7, 8, 9], presumably as this mimics the in vivo lipid primer. The enzymes have been used in applications to modify therapeutic proteins and prepare synthetic vaccines [10, 11], where un-natural acceptors like protein N-glycans have been used.
Kinetics and Mechanism
Sialic acid transfer occurs with inversion of configuration (from the β-linked CMP-Neu5Ac donor to the α-2,8-linked polySia), and polyST has been proposed to follow a SN2-like direct displacement mechanism. While H291 could act as a catalytic acid to stabilize the nucleotide phosphate-leaving group.
Catalytic Residues
Residues involved in catalysis have been proposed from site-directed mutagensis and the X-ray crystal structure from the M. hemolytica serotype A2 enzyme [12]. The catalytic base E153 abstracts a proton from the C8′ hydroxyl group of the sialic acid acceptor concerted with the nucleophilic attack on the anomeric C2′ carbon of the CMP-sialic acid donor substrate, thereby generating an α-2,8 glycosidic linkage. The resulting negatively charged CMP leaving group is stabilized by H291 assisted by S339 and T340.
Three-dimensional structures
- PDB ID 5WC6 from GT38 (click images for large versions)
The structure is a GT-B fold typical of a metal independent glycosyltransferase, it has 2 non-equivalent Rossmann-like folds. The structure was solved from a truncated version of the enzyme, which lacks 20 amino acids from the N-terminal end. There are two protomers in the crystal structure, but biochemical evidence suggests the soluble enzyme exists as a monomer. There is a hinge region between these domains (F227 to N236) which gives some flexibility in the structure. The structure shows an N-terminal tail which is unstructured and is likely to be a linker to the membrane anchor. The structure has a large electropositive groove which accomodates the polySia chain (right hand panel). One of the additional structures obtained for this enzyme is a complex with the synthetic heparin fondaparinux which was a surrogate for the polyanionic polySia. The image shown here is the CDP donor analogue complex which sites on one the Rossmann like domains.
Family Firsts
- First general acid/base residue identification
- E153, H291
Prior to having the structure, it had been suggested that the motif "HP" was involved in catalysis. Site directed mutagenesis with the MhpolyST suggested that the H291 residue does play an essential role in catalysis - as the general acid. This "HP" motif is also conserved in other bacterial sialytransferases. Similarly, the E153 residue has been proposed as the general base for this reaction. Both E153 and H291 show large decreases in kcat/Km when mutated to alanine - consistent with that assessment.
- First 3-D structure
- The family of structures are: PDB codes 5WC8, 5WCN, 5WC6 and 5WD7.
There are a family of structures, an apo-structure 5WC8, an acceptor complex (Sia2LacNAc6S) 5WCN, a donor analogue (CDP)5WC6, and a product analogue complex (fondapariux) 5WD7.
All of these structures required a mutation to decrease surface entropy - K69A.
References
- Cho JW and Troy FA 2nd. (1994). Polysialic acid engineering: synthesis of polysialylated neoglycosphingolipids by using the polysialyltransferase from neuroinvasive Escherichia coli K1. Proc Natl Acad Sci U S A. 1994;91(24):11427-31. DOI:10.1073/pnas.91.24.11427 |
- Willis LM, Stupak J, Richards MR, Lowary TL, Li J, and Whitfield C. (2013). Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporter-dependent pathways in Gram-negative pathogens. Proc Natl Acad Sci U S A. 2013;110(19):7868-73. DOI:10.1073/pnas.1222317110 |
- Jennings HJ, Bhattacharjee AK, Bundle DR, Kenny CP, Martin A, and Smith IC. (1977). Strucutres of the capsular polysaccharides of Neisseria meningitidis as determined by 13C-nuclear magnetic resonance spectroscopy. J Infect Dis. 1977;136 Suppl:S78-83. DOI:10.1093/infdis/136.supplement.s78 |
- Puente-Polledo L, Reglero A, González-Clemente C, Rodríguez-Aparicio LB, and Ferrero MA. (1998). Biochemical conditions for the production of polysialic acid by Pasteurella haemolytica A2. Glycoconj J. 1998;15(9):855-61. DOI:10.1023/a:1006902931032 |
- Devi SJ, Schneerson R, Egan W, Vann WF, Robbins JB, and Shiloach J. (1991). Identity between polysaccharide antigens of Moraxella nonliquefaciens, group B Neisseria meningitidis, and Escherichia coli K1 (non-O acetylated). Infect Immun. 1991;59(2):732-6. DOI:10.1128/iai.59.2.732-736.1991 |
- Glode MP, Robbins JB, Liu TY, Gotschlich EC, Orskov I, and Orskov F. (1977). Cross-antigenicity and immunogenicity between capsular polysaccharides of group C Neisseria meningitidis and of Escherichia coli K92. J Infect Dis. 1977;135(1):94-104. DOI:10.1093/infdis/135.1.94 |
- Willis LM, Gilbert M, Karwaski MF, Blanchard MC, and Wakarchuk WW. (2008). Characterization of the alpha-2,8-polysialyltransferase from Neisseria meningitidis with synthetic acceptors, and the development of a self-priming polysialyltransferase fusion enzyme. Glycobiology. 2008;18(2):177-86. DOI:10.1093/glycob/cwm126 |
- Peterson DC, Arakere G, Vionnet J, McCarthy PC, and Vann WF. (2011). Characterization and acceptor preference of a soluble meningococcal group C polysialyltransferase. J Bacteriol. 2011;193(7):1576-82. DOI:10.1128/JB.00924-10 |
- Lindhout T, Bainbridge CR, Costain WJ, Gilbert M, and Wakarchuk WW. (2013). Biochemical characterization of a polysialyltransferase from Mannheimia haemolytica A2 and comparison to other bacterial polysialyltransferases. PLoS One. 2013;8(7):e69888. DOI:10.1371/journal.pone.0069888 |
- Lindhout T, Iqbal U, Willis LM, Reid AN, Li J, Liu X, Moreno M, and Wakarchuk WW. (2011). Site-specific enzymatic polysialylation of therapeutic proteins using bacterial enzymes. Proc Natl Acad Sci U S A. 2011;108(18):7397-402. DOI:10.1073/pnas.1019266108 |
- McCarthy PC, Saksena R, Peterson DC, Lee CH, An Y, Cipollo JF, and Vann WF. (2013). Chemoenzymatic synthesis of immunogenic meningococcal group C polysialic acid-tetanus Hc fragment glycoconjugates. Glycoconj J. 2013;30(9):857-70. DOI:10.1007/s10719-013-9490-x |
- Lizak C, Worrall LJ, Baumann L, Pfleiderer MM, Volkers G, Sun T, Sim L, Wakarchuk W, Withers SG, and Strynadka NCJ. (2017). X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid. Sci Rep. 2017;7(1):5842. DOI:10.1038/s41598-017-05627-z |