CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "General acid/base"
(Created page with ' * Author: Spencer Williams * Responsible Curator: Spencer Williams ---- == Overview == The term '''catalytic acid/base''' re…') |
|||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | {{CuratorApproved}} | |
* Author: [[User:SpencerWilliams|Spencer Williams]] | * Author: [[User:SpencerWilliams|Spencer Williams]] | ||
| Line 6: | Line 6: | ||
== Overview == | == Overview == | ||
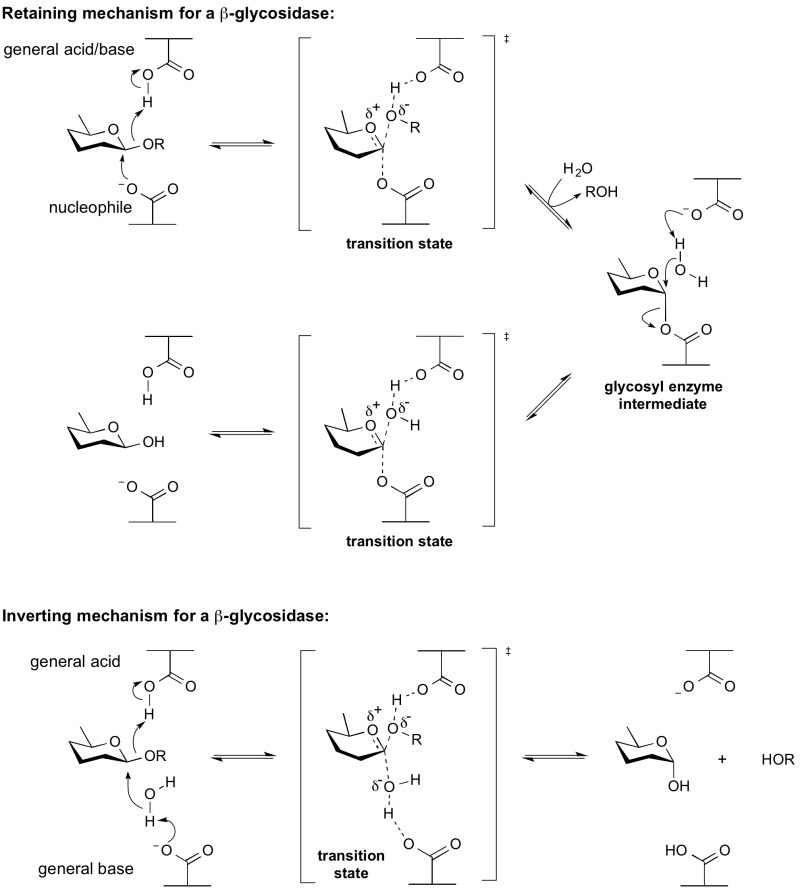
| − | The term ''' | + | The term '''general acid/base''' (also catalytic acid/base) refers to an amino acid residue in a [[glycoside hydrolase]] or a related enzyme that participates in the mechanism of hydrolysis by removing or adding a proton (or both). The mechanism may be a [[retaining]] or [[inverting]] mechanism. General acid/base catalysis differs from specific acid/base catalysis as in the latter it is the solvent that acts as the acid or base. |
| + | |||
| + | [[Image:General_acid_base.png|centre|800px]] | ||
| + | |||
| + | ==Methods for indentifying the general acid/base== | ||
| + | |||
| + | ===Kinetic analysis of mutants=== | ||
| − | [[ | + | Assignment of the acid/base residues may be achieved by the kinetic analysis of enzyme mutants in which the candidate residues have been replaced by a non-nucleophilic residue. Alanine is preferred as a replacement residue owing to its small size. Glycine is non-optimal, as the lack of an alpha-substituent may lead to differences in conformation. Alanine mutants typically have ''k''<sub>cat</sub> values 1,000-fold lower than the wildtype enzyme when assessed against the natural substrate <cite>Ly1999</cite>.This effect is much smaller when the substrate possesses a good leaving group, such as a fluoride or a 2,4-dinitrophenyl group, as in these cases the leaving groups do not require general acid catalysis. In these cases the ''k''<sub>cat</sub> values are unaffected, but as the deglycosylation step requires general base catalysis the [[intermediate]] accumulates, leading to a reduction in the ''K''<sub>M</sub> value. A further test for the identity of the acid base residue is the observation of a 'chemical rescue' effect. In the presence of a small anionic nucleophilic group such as azide, fluoride or formate, the glycosyl enzyme will be turned over. In this case the product of 'chemical rescue' should be a glycosyl azide, fluoride or formate with anomeric stereochemistry matching that of the starting glycoside. |
| − | == | + | ===Mechanism-based labelling=== |
| + | |||
| + | Various electrophilic reagents including ''N''-bromoacetyl glycosylamines, epoxyalkyl glycosides, and alpha-halo ketones have been used with some success to identify the general acid/base residue. However, such labelling studies are not entirely reliable, and candidate acid/base residues identified through their use should be regarded as preliminary assignments and follow up experiments involving kinetic analysis of mutants should be performed for improving the confidence of the assignment. | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | + | #Ly1999 pmid=10872458 | |
</biblio> | </biblio> | ||
[[Category:Definitions and explanations]] | [[Category:Definitions and explanations]] | ||
Latest revision as of 02:38, 4 August 2013
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: Spencer Williams
- Responsible Curator: Spencer Williams
Overview
The term general acid/base (also catalytic acid/base) refers to an amino acid residue in a glycoside hydrolase or a related enzyme that participates in the mechanism of hydrolysis by removing or adding a proton (or both). The mechanism may be a retaining or inverting mechanism. General acid/base catalysis differs from specific acid/base catalysis as in the latter it is the solvent that acts as the acid or base.
Methods for indentifying the general acid/base
Kinetic analysis of mutants
Assignment of the acid/base residues may be achieved by the kinetic analysis of enzyme mutants in which the candidate residues have been replaced by a non-nucleophilic residue. Alanine is preferred as a replacement residue owing to its small size. Glycine is non-optimal, as the lack of an alpha-substituent may lead to differences in conformation. Alanine mutants typically have kcat values 1,000-fold lower than the wildtype enzyme when assessed against the natural substrate [1].This effect is much smaller when the substrate possesses a good leaving group, such as a fluoride or a 2,4-dinitrophenyl group, as in these cases the leaving groups do not require general acid catalysis. In these cases the kcat values are unaffected, but as the deglycosylation step requires general base catalysis the intermediate accumulates, leading to a reduction in the KM value. A further test for the identity of the acid base residue is the observation of a 'chemical rescue' effect. In the presence of a small anionic nucleophilic group such as azide, fluoride or formate, the glycosyl enzyme will be turned over. In this case the product of 'chemical rescue' should be a glycosyl azide, fluoride or formate with anomeric stereochemistry matching that of the starting glycoside.
Mechanism-based labelling
Various electrophilic reagents including N-bromoacetyl glycosylamines, epoxyalkyl glycosides, and alpha-halo ketones have been used with some success to identify the general acid/base residue. However, such labelling studies are not entirely reliable, and candidate acid/base residues identified through their use should be regarded as preliminary assignments and follow up experiments involving kinetic analysis of mutants should be performed for improving the confidence of the assignment.