CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 106"
(Created page with " <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> {{UnderConstruc...") |
Wenwen Tao (talk | contribs) |
||
| (11 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
* [[Author]]: [[User:Wenwen Tao|Wenwen Tao]] | * [[Author]]: [[User:Wenwen Tao|Wenwen Tao]] | ||
* [[Responsible Curator]]: [[User:Yaoguang Chang|Yaoguang Chang]] | * [[Responsible Curator]]: [[User:Yaoguang Chang|Yaoguang Chang]] | ||
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | + | VbCBM106 represents the first characterized member of the CBM106 family, which is appended to a [[PL6]] potential alginate lyase (GenBank: ANO33061.1) from the marine bacterium ''Vibrio breoganii'' <cite>Hidalgo2009</cite>. The CBM showed the favorable specificity to alginate, while it could not bind to other polyuronic acids, such as hyaluronic acid, chondroitin sulfates, dermatan sulfate, and pectin, as well as other polysaccharides from brown algae including laminarin and fucoidan <cite>Mei2024</cite>. | |
| − | |||
| − | '' | ||
== Structural Features == | == Structural Features == | ||
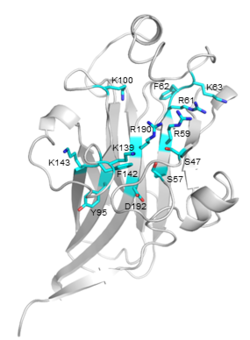
| − | + | [[File:CBM106_Fig.1.png|thumb|250px|right|'''Figure 1. Crystal structure of VbCBM106 ([{{PDBlink}}8zqf PDB 8zqf]).''' The strand, helix, and loop were colored in cyan, yellow, and white respectively. The sequential order of these strands and helices was depicted from P4 through to G200.]] | |
| − | + | [[File:CBM106_Fig.2.png|thumb|250px|right|'''Figure 2. Detailed view of the predicted binding site of VbCBM106.''' The potential critical residues and their side chains are shown.]] | |
| − | + | The crystal structure (1.55 Å) of VbCBM106 exhibits a typical β-sandwich fold, which is composed of two antiparallel β-sheets formed by 11 β-strands and 4 helixes (Fig.1). Several conserved residues were identified, including S47, S57, R59, R61, F62, K63, Y95, K100, K139, F142, K143, R190, and D192. The conserved residues are spatially adjacent, situated on the concave of one β-sheet and the apical loop region (Fig.2). Site-directed mutagenesis assays demonstrated that the residues R59, R61, K63, K139, and R190 play critical roles in the ligand binding of VbCBM106 <cite>Mei2024</cite>. | |
| − | + | Since VbCBM106 is capable of binding to soluble alginate polysaccharides, it is suggested that it belongs to type B <cite>Boraston2004</cite>. | |
| − | |||
== Functionalities == | == Functionalities == | ||
| − | + | VbCBM106 and some of its homologous sequences are linked to the catalytic modules of the PL6 family or its subfamily PL6_1. In the natural environments, VbCBM106 might enhance the catalytic efficiency of its appended enzymes by increasing the contact between adjacent catalytic domain and alginate, similar to the role of the CBM13 domain in alginate lyase <cite>Li2015</cite>. | |
| − | |||
| − | |||
| − | |||
== Family Firsts == | == Family Firsts == | ||
| − | ;First Identified | + | ;First Identified: The first member VbCBM106 is a component of a potential [[PL6]] alginate lyase from a marine bacterium ''Vibrio breoganii'' <cite>Mei2024</cite>. |
| − | : | + | ;First Structural Characterization: VbCBM106 ([{{PDBlink}}8zqf PDB 8zqf]) is the first and currently only member of the CBM106 family with the structural information <cite>Mei2024</cite>. |
| − | ;First Structural Characterization | ||
| − | : | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Hidalgo2009 pmid=19542146 |
| − | # | + | #Mei2024 pmid=39069041 |
#Boraston2004 pmid=15214846 | #Boraston2004 pmid=15214846 | ||
| − | # | + | #Li2015 pmid=25837818 |
| − | |||
| − | |||
| − | |||
</biblio> | </biblio> | ||
Latest revision as of 22:06, 20 November 2024
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM106.html |
Ligand specificities
VbCBM106 represents the first characterized member of the CBM106 family, which is appended to a PL6 potential alginate lyase (GenBank: ANO33061.1) from the marine bacterium Vibrio breoganii [1]. The CBM showed the favorable specificity to alginate, while it could not bind to other polyuronic acids, such as hyaluronic acid, chondroitin sulfates, dermatan sulfate, and pectin, as well as other polysaccharides from brown algae including laminarin and fucoidan [2].
Structural Features

The crystal structure (1.55 Å) of VbCBM106 exhibits a typical β-sandwich fold, which is composed of two antiparallel β-sheets formed by 11 β-strands and 4 helixes (Fig.1). Several conserved residues were identified, including S47, S57, R59, R61, F62, K63, Y95, K100, K139, F142, K143, R190, and D192. The conserved residues are spatially adjacent, situated on the concave of one β-sheet and the apical loop region (Fig.2). Site-directed mutagenesis assays demonstrated that the residues R59, R61, K63, K139, and R190 play critical roles in the ligand binding of VbCBM106 [2]. Since VbCBM106 is capable of binding to soluble alginate polysaccharides, it is suggested that it belongs to type B [3].
Functionalities
VbCBM106 and some of its homologous sequences are linked to the catalytic modules of the PL6 family or its subfamily PL6_1. In the natural environments, VbCBM106 might enhance the catalytic efficiency of its appended enzymes by increasing the contact between adjacent catalytic domain and alginate, similar to the role of the CBM13 domain in alginate lyase [4].
Family Firsts
- First Identified
- The first member VbCBM106 is a component of a potential PL6 alginate lyase from a marine bacterium Vibrio breoganii [2].
- First Structural Characterization
- VbCBM106 (PDB 8zqf) is the first and currently only member of the CBM106 family with the structural information [2].
References
- Beaz Hidalgo R, Cleenwerck I, Balboa S, Prado S, De Vos P, and Romalde JL. (2009). Vibrio breoganii sp. nov., a non-motile, alginolytic, marine bacterium within the Vibrio halioticoli clade. Int J Syst Evol Microbiol. 2009;59(Pt 7):1589-94. DOI:10.1099/ijs.0.003434-0 |
- Mei X, Tao W, Sun H, Liu G, Chen G, Zhang Y, Xue C, and Chang Y. (2024). Characterization and structural identification of a novel alginate-specific carbohydrate-binding module (CBM): The founding member of a new CBM family. Int J Biol Macromol. 2024;277(Pt 3):134221. DOI:10.1016/j.ijbiomac.2024.134221 |
- Boraston AB, Bolam DN, Gilbert HJ, and Davies GJ. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769-81. DOI:10.1042/BJ20040892 |
- Li S, Yang X, Bao M, Wu Y, Yu W, and Han F. (2015). Family 13 carbohydrate-binding module of alginate lyase from Agarivorans sp. L11 enhances its catalytic efficiency and thermostability, and alters its substrate preference and product distribution. FEMS Microbiol Lett. 2015;362(10). DOI:10.1093/femsle/fnv054 |