CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 105"
Guanchen Liu (talk | contribs) |
|||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | The first member of family CBM105 ( | + | The first member of family CBM105 (SoCBM105) was identified in the [[PL29]] multidomain chondroitinase SoChABC29 from ''Segatella oris'' <cite>Liu2024</cite>. SoCBM105 bound specifically to chondroitin sulfates (CSs) including CS-A and CS-C, while it was incapable of binding to other glycosaminoglycans or polyuronic acid substrates <cite>Liu2024</cite>. |
== Structural Features == | == Structural Features == | ||
| − | [[File:CBM105 Fig.1.png|thumb|350px|right|'''Figure 1. Domain analysis of | + | [[File:CBM105 Fig.1.png|thumb|350px|right|'''Figure 1. Domain analysis of SoChABC29, the parent enzyme of SoCBM105 <cite>Jumper2021</cite>.''' The enzyme consists of a signal peptide (1-21 amino acids), a PL29 domain (66-380 amino acids) and a CBM105 domain (viz., SoCBM105; 555-785 amino acids).]] |
| − | An AlphaFold2 model predicts that SoCBM105 has a β-sandwich fold (Fig.1). | + | An AlphaFold2 <cite>Jumper2021</cite> model predicts that SoCBM105 has a β-sandwich fold (Fig.1). |
== Functionalities == | == Functionalities == | ||
| − | + | SoCBM105 is at the C-terminus domain of a [[PL29]] enzyme SoChABC29 that displays chondroitin sulfate ABC activity, consistent with the SoCBM105 specificity <cite>Liu2024</cite>. Biochemical characterization of SoChABC29 and the CBM-truncated enzyme revealed that the SoCBM105 enhances the catalytic activity, thermostability, and disaccharide proportion in the final enzymatic products of SoChABC29 <cite>Liu2024</cite>. | |
== Family Firsts == | == Family Firsts == | ||
| − | ;First Identified | + | ;First Identified: Binding to chondroitin sulfate in the CBM105 family was first characterized and identified for SoCBM105 from the ''S. oris'' [[PL29]] chondroitinase <cite>Liu2024</cite>. |
| − | : | + | ;First Structural Characterization: No experimentally determined three-dimensional structure has been solved in this CBM family. |
| − | ;First Structural Characterization | ||
| − | :No experimentally determined three-dimensional structure has been solved in this CBM family. | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
#Liu2024 pmid=38777025 | #Liu2024 pmid=38777025 | ||
| + | #Jumper2021 pmid=34265844 | ||
</biblio> | </biblio> | ||
Latest revision as of 20:33, 6 November 2024
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM105.html |
Ligand specificities
The first member of family CBM105 (SoCBM105) was identified in the PL29 multidomain chondroitinase SoChABC29 from Segatella oris [1]. SoCBM105 bound specifically to chondroitin sulfates (CSs) including CS-A and CS-C, while it was incapable of binding to other glycosaminoglycans or polyuronic acid substrates [1].
Structural Features
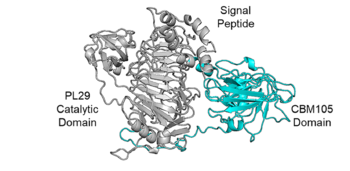
An AlphaFold2 [2] model predicts that SoCBM105 has a β-sandwich fold (Fig.1).
Functionalities
SoCBM105 is at the C-terminus domain of a PL29 enzyme SoChABC29 that displays chondroitin sulfate ABC activity, consistent with the SoCBM105 specificity [1]. Biochemical characterization of SoChABC29 and the CBM-truncated enzyme revealed that the SoCBM105 enhances the catalytic activity, thermostability, and disaccharide proportion in the final enzymatic products of SoChABC29 [1].
Family Firsts
- First Identified
- Binding to chondroitin sulfate in the CBM105 family was first characterized and identified for SoCBM105 from the S. oris PL29 chondroitinase [1].
- First Structural Characterization
- No experimentally determined three-dimensional structure has been solved in this CBM family.
References
- Liu G, Song L, Li J, Song X, Mei X, Zhang Y, Fan C, Chang Y, and Xue C. (2024). Identification and characterization of a chondroitinase ABC with a novel carbohydrate-binding module. Int J Biol Macromol. 2024;271(Pt 1):132518. DOI:10.1016/j.ijbiomac.2024.132518 |
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, and Hassabis D. (2021). Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583-589. DOI:10.1038/s41586-021-03819-2 |