CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 96"
Wenwen Tao (talk | contribs) |
Wenwen Tao (talk | contribs) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | Members of the CBM96 family are present within various CAZyme families, including [[PL8]], [[PL35]], [[GH16]], and [[GH136]], suggesting that they may possess a wide range of diverse activities. TM6-N4, originating from the marine thermophilic bacterium ''Defluviitalea phaphyphila'', and DmCBM96-1/DmCBM96-2, derived from the bacterium ''Dysgonomonas sp.'' HDW5A isolated from ''Hydrophilus acuminatus'' , are currently the characterized members in the CBM96 family. TM6-N4 is associated with the catalytic module of [[PL39]] alginate lyase, and its specific binding capacity to alginate was confirmed by affinity electrophoresis analysis. Isothermal titration calorimetry analysis revealed that TM6-N4 exhibits no significant preference for mannuronate or guluronate oligosaccharides <cite>Ji2023</cite>. Compared with TM6-N4, DmCBM96-1 exhibits a relatively low sequence identity of approximately 32%, and DmCBM96-2 is even lower of approximately 25%. DmCBM96-1 and DmCBM96-2, which are consecutive CBMs linked to chondroitin sulfate (CS) lyase | + | Members of the CBM96 family are present within various CAZyme families, including [[PL8]], [[PL35]], [[GH16]], and [[GH136]], suggesting that they may possess a wide range of diverse activities. TM6-N4, originating from the marine thermophilic bacterium ''Defluviitalea phaphyphila'', and DmCBM96-1/DmCBM96-2, derived from the bacterium ''Dysgonomonas sp.'' HDW5A isolated from ''Hydrophilus acuminatus'' , are currently the characterized members in the CBM96 family. TM6-N4 is associated with the catalytic module of [[PL39]] alginate lyase, and its specific binding capacity to alginate was confirmed by affinity electrophoresis analysis. Isothermal titration calorimetry analysis revealed that TM6-N4 exhibits no significant preference for mannuronate or guluronate oligosaccharides <cite>Ji2023</cite>. Compared with TM6-N4, DmCBM96-1 exhibits a relatively low sequence identity of approximately 32%, and DmCBM96-2 is even lower of approximately 25% <cite>Altschul1997</cite>. DmCBM96-1 and DmCBM96-2, which are consecutive CBMs linked to [[PL8]]_3 chondroitin sulfate (CS) lyase, have been confirmed to bind to CS-A (chondroitin 4-sulfate), CS-C (chondroitin 6-sulfate), and Aj-fCS (consisting of CS-type backbone and sulfated fucose branches), but not to other polyuronic acids such as hyaluronic acid, dermatan sulfate, heparin, pectin, and alginate. Comparatively, the relative affinity of both DmCBM96 proteins to CS-C are higher than CS-A <cite>Liu2024</cite>. |
== Structural Features == | == Structural Features == | ||
| − | [[File:CBM96_Fig.1.png|thumb|350px|right|'''Figure 1. Crystal structure of TM6-N4.''' The strand, helix, loop and Ca<Sup>2+</Sup> were colored in cyan, yellow, white and green respectively.]] | + | [[File:CBM96_Fig.1.png|thumb|350px|right|'''Figure 1. Crystal structure of TM6-N4 ([{{PDBlink}}7vbo PDB 7vbo]).''' The strand, helix, loop and Ca<Sup>2+</Sup> were colored in cyan, yellow, white and green respectively.]] |
| − | The crystal structures of TM6-N4 (1.35 Å) and DmCBM96-1 (2.20 Å) reveal that they both exhibit a typical β-sandwich fold, comprising two antiparallel β-sheets consisting of 11 β-strands, but differ in helixes. The binding sites of both DmCBM96-1 and TM6-N4 are located within the loop regions. Site-directed mutagenesis assay revealed that the basic amino acids Arg27, Lys45, Arg53, Arg157, and the aromatic amino acid Tyr51 at the binding site of DmCBM96-1 are crucial for CS binding <cite>Liu2024</cite>. While , the amino acids located at the bottom or wall of the shallow groove of TM6-N4, including Lys10, Lys22, Lys25, Lys27, Lys31, Arg36, and Tyr159, are essential for alginate binding <cite>Ji2023</cite>. Among these, the crucial amino acids Lys45 and Arg53 in DmCBM96-1 correspond to the key amino acids Lys27 and Arg36 in TM6-N4, respectively, and they are highly conserved within the CBM96 family, suggesting their pivotal roles in the ligand binding process. | + | The crystal structures of TM6-N4 (1.35 Å) and DmCBM96-1 (2.20 Å) reveal that they both exhibit a typical β-sandwich fold, comprising two antiparallel β-sheets consisting of 11 β-strands, but differ in helixes. The binding sites of both DmCBM96-1 and TM6-N4 are located within the loop regions. Site-directed mutagenesis assay revealed that the basic amino acids Arg27, Lys45, Arg53, Arg157, and the aromatic amino acid Tyr51 at the binding site of DmCBM96-1 are crucial for CS binding <cite>Liu2024</cite>. While , the amino acids located at the bottom or wall of the shallow groove of TM6-N4, including Lys10, Lys22, Lys25, Lys27, Lys31, Arg36, and Tyr159, are essential for alginate binding <cite>Ji2023</cite>. Among these, the crucial amino acids Lys45 and Arg53 in DmCBM96-1 correspond to the key amino acids Lys27 and Arg36 in TM6-N4, respectively, and they are highly conserved within the CBM96 family, suggesting their pivotal roles in the ligand binding process. Structural and mutational analyses revealed that TM6-N4 possess long-extended binding grooves for alginate chains, characteristic of type B CBM fold <cite>Ji2023</cite>. |
== Functionalities == | == Functionalities == | ||
| − | Members of the CBM96 family are annotated by dbCAN to be associated with several CAZyme families, and their presumed physiological role is to facilitate enzyme-carbohydrate interactions and enhance the local concentration of enzymes near substrates, thereby promoting the degradation of polysaccharides. Based on the polysaccharide-binding specificity of the CBM96 family, it may also have potential applications in the study of cellular structures or the detection of pathogenic bacteria. Furthermore, through fusion expression, CBM96 could potentially be utilized to optimize the properties of other CAZymes <cite>Ji2023</cite>. | + | Members of the CBM96 family are annotated by dbCAN to be associated with several CAZyme families <cite>Yin2012</cite>, and their presumed physiological role is to facilitate enzyme-carbohydrate interactions and enhance the local concentration of enzymes near substrates, thereby promoting the degradation of polysaccharides. Based on the polysaccharide-binding specificity of the CBM96 family, it may also have potential applications in the study of cellular structures or the detection of pathogenic bacteria. Furthermore, through fusion expression, CBM96 could potentially be utilized to optimize the properties of other CAZymes <cite>Ji2023</cite>. |
== Family Firsts == | == Family Firsts == | ||
| Line 35: | Line 35: | ||
#Ji2023 pmid=36592931 | #Ji2023 pmid=36592931 | ||
#Liu2024 pmid=38805590 | #Liu2024 pmid=38805590 | ||
| + | #Yin2012 pmid=22645317 | ||
| + | #Altschul1997 pmid=9254694 | ||
</biblio> | </biblio> | ||
Latest revision as of 22:05, 20 November 2024
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM96.html |
Ligand specificities
Members of the CBM96 family are present within various CAZyme families, including PL8, PL35, GH16, and GH136, suggesting that they may possess a wide range of diverse activities. TM6-N4, originating from the marine thermophilic bacterium Defluviitalea phaphyphila, and DmCBM96-1/DmCBM96-2, derived from the bacterium Dysgonomonas sp. HDW5A isolated from Hydrophilus acuminatus , are currently the characterized members in the CBM96 family. TM6-N4 is associated with the catalytic module of PL39 alginate lyase, and its specific binding capacity to alginate was confirmed by affinity electrophoresis analysis. Isothermal titration calorimetry analysis revealed that TM6-N4 exhibits no significant preference for mannuronate or guluronate oligosaccharides [1]. Compared with TM6-N4, DmCBM96-1 exhibits a relatively low sequence identity of approximately 32%, and DmCBM96-2 is even lower of approximately 25% [2]. DmCBM96-1 and DmCBM96-2, which are consecutive CBMs linked to PL8_3 chondroitin sulfate (CS) lyase, have been confirmed to bind to CS-A (chondroitin 4-sulfate), CS-C (chondroitin 6-sulfate), and Aj-fCS (consisting of CS-type backbone and sulfated fucose branches), but not to other polyuronic acids such as hyaluronic acid, dermatan sulfate, heparin, pectin, and alginate. Comparatively, the relative affinity of both DmCBM96 proteins to CS-C are higher than CS-A [3].
Structural Features
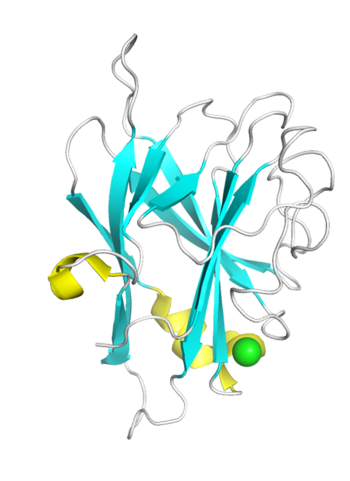
The crystal structures of TM6-N4 (1.35 Å) and DmCBM96-1 (2.20 Å) reveal that they both exhibit a typical β-sandwich fold, comprising two antiparallel β-sheets consisting of 11 β-strands, but differ in helixes. The binding sites of both DmCBM96-1 and TM6-N4 are located within the loop regions. Site-directed mutagenesis assay revealed that the basic amino acids Arg27, Lys45, Arg53, Arg157, and the aromatic amino acid Tyr51 at the binding site of DmCBM96-1 are crucial for CS binding [3]. While , the amino acids located at the bottom or wall of the shallow groove of TM6-N4, including Lys10, Lys22, Lys25, Lys27, Lys31, Arg36, and Tyr159, are essential for alginate binding [1]. Among these, the crucial amino acids Lys45 and Arg53 in DmCBM96-1 correspond to the key amino acids Lys27 and Arg36 in TM6-N4, respectively, and they are highly conserved within the CBM96 family, suggesting their pivotal roles in the ligand binding process. Structural and mutational analyses revealed that TM6-N4 possess long-extended binding grooves for alginate chains, characteristic of type B CBM fold [1].
Functionalities
Members of the CBM96 family are annotated by dbCAN to be associated with several CAZyme families [4], and their presumed physiological role is to facilitate enzyme-carbohydrate interactions and enhance the local concentration of enzymes near substrates, thereby promoting the degradation of polysaccharides. Based on the polysaccharide-binding specificity of the CBM96 family, it may also have potential applications in the study of cellular structures or the detection of pathogenic bacteria. Furthermore, through fusion expression, CBM96 could potentially be utilized to optimize the properties of other CAZymes [1].
Family Firsts
- First Identified
- The TM6-N4 derived from the alginate lyase Dp0100 of marine bacterium Defluviitalea phaphyphila, which was first reported in 2023, represents the first characterized member of the CBM96 family [1].
- First Structural Characterization
- Alginate-binding protein TM6-N4 is the first member of the CBM96 family to undergo X-ray crystallography structural characterization (PDB 7vbo) [1]. Subsequently, CS-binding protein DmCBM96-1 was also characterized (PDB 2pqr) [3].
References
- Ji S, Tian X, Li X, and She Q. (2023). Identification and structural analysis of a carbohydrate-binding module specific to alginate, a representative of a new family, CBM96. J Biol Chem. 2023;299(2):102854. DOI:10.1016/j.jbc.2022.102854 |
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389-402. DOI:10.1093/nar/25.17.3389 |
- Liu G, Mei X, Zhang Y, Chen G, Li J, Tao W, Sun M, Zheng L, Chang Y, and Xue C. (2024). Characterization and Structural Analysis of a Novel Carbohydrate-Binding Module from Family 96 with Chondroitin Sulfate-Specific Binding Capacity. J Agric Food Chem. 2024;72(23):13196-13204. DOI:10.1021/acs.jafc.4c00090 |
- Yin Y, Mao X, Yang J, Chen X, Mao F, and Xu Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40(Web Server issue):W445-51. DOI:10.1093/nar/gks479 |