CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 58"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (29 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <!-- CURATORS: Please | + | <!-- CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> |
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Warren Wakarchuk|Warren Wakarchuk]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Warren Wakarchuk|Warren Wakarchuk]] |
---- | ---- | ||
| Line 18: | Line 18: | ||
|- | |- | ||
|'''Active site residues''' | |'''Active site residues''' | ||
| − | | | + | |Arg596, Arg647 and Glu581 |
|- | |- | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
|- | |- | ||
| − | | colspan="2" | | + | | colspan="2" |{{CAZyDBlink}}GH58.html |
|} | |} | ||
</div> | </div> | ||
| Line 29: | Line 29: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | [[Glycoside hydrolases]] of family 58 are [[endo]]-''N''-acetylneuraminidases (also termed [[endo]]-sialidases). The capsular coat surrounding ''E. coli'' K1, protects the bacterium from | + | [[Glycoside hydrolases]] of family 58 are [[endo]]-''N''-acetylneuraminidases (also termed [[endo]]-sialidases). The capsular coat surrounding ''E. coli'' K1, is poly-α-2,8-sialic acid and protects the bacterium from the host immune system, but it also acts as an anchor point for bacteriophage infection. A range of bacteriophages specific to this ''E. coli'' capsule type have been described which have tailspike enzymes that degrade the capuslar material <cite>Kwiatowski Hallenbeck Petter</cite> |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | + | Family 58 [[endo]]-sialidases are specific for polysialic acid and form a unique family of [[endo]]-acting sialidases, all others previously reported being [[exo]]-acting. The recently determined structure of an [[endo]]-sialidase derived from bacteriophage K1F (endoNF) <cite>Stummeyer</cite> revealed the active site to lack a number of the residues that are conserved in other sialidases, implying a new, [[endo]]-sialidase-specific catalytic mechanism. Using synthetic trifluoromethylumbelliferyl oligosialoside substrates kinetic parameters for hydrolysis at a single cleavage site were determined. Measurement of ''k<sub>cat</sub>/K<sub>m</sub>'' at a series of pH values revealed a dependence on a single protonated group of pK<sub>a</sub> 5. Direct <sup>1</sup>H-NMR analysis of the hydrolysis of trifluoromethylumbelliferyl sialotrioside revealed that endoNF affects catalysis with an [[inverting]] mechanism <cite>Morely</cite>. | |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | + | From the crystal structure of the GH-58 enzyme it was noted that the normal catalyic residues found in exo-sialidases were not present. Instead, three conserved amino acids (Arg596, Arg647 and Glu581) were observed that contribute to endosialidase activity. Single mutations of those residues reduced the level of activity, but not enough to mark the residues as absolutely required. However, when double mutants were examined there was a complete loss of hydrolysis, but binding to PSA still occured <cite>Stummeyer</cite>. Since this enzyme proceeds with inverting stereochemistry, a catalytic base is implied by the mechanism, and Glu581 might fill that role <cite>Morely</cite>. | |
| − | |||
== Three-dimensional structures == | == Three-dimensional structures == | ||
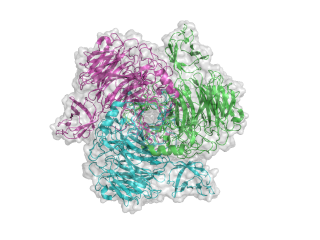
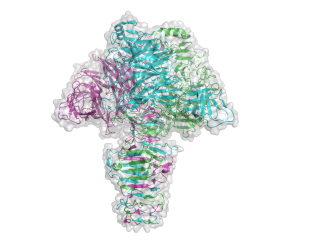
| − | The structure for the K1F tailspike enzyme has been solved by Stummeyer et al. <cite> | + | The structure for the K1F tailspike enzyme has been solved by Stummeyer et al. <cite>Stummeyer</cite>. The protein structure is similar to other phage tailspike endo-hydrolases in that it is an extended, trimeric molecule. The first tailspike enzyme to be studied in detail was the endo-rhamnosidase from the Salmonella phage P22 <cite>Steinbacher</cite>, which is in family [[GH90]]. The pictures here were generated in [http://www.pymol.org/ Pymol] from PDB entry [{{PDBlink}}1voe 1voe]. Each monomer is a different colour to highlight the trimeric nature of the protein. |
| − | + | <gallery widths=320px heights=240px perrow=2 caption="PDB ID 1voe from GH58 (click images for large versions)"> | |
| + | File:Gh581.png|top view | ||
| + | File:Gh58_2.png|side view | ||
| + | </gallery> | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First sterochemistry determination: The stereochemical outcome of this reaction was determined in the Withers laboratory at UBC using synthetic substrates with a trifluoromethyl-umbelliferol fluorophore <cite> | + | ;First sterochemistry determination: The stereochemical outcome of this reaction was determined in the Withers laboratory at UBC using synthetic substrates with a trifluoromethyl-umbelliferol fluorophore <cite>Morely</cite>. This determination showed that unlike all other known sialidases, the [[endo]]-sialidase uses an [[inverting]] mechanism. |
| − | ;First [[catalytic nucleophile]] identification: | + | ;First [[catalytic nucleophile]] identification: Since this enzyme proceeds with inversion of sterechemistry, is does not use a catalytic nuceleophile. |
| − | ;First [[general acid/base]] residue identification: | + | ;First [[general acid/base]] residue identification: It has been suggested though not conclusively proven that Glu581 is the catalytic base for this reaction <cite>Stummeyer Morely</cite>. |
| − | ;First 3-D structure: The first structure determination was performed with the K1F protein <cite> | + | ;First 3-D structure: The first structure determination was performed with a truncated version of the K1F protein <cite>Stummeyer</cite>. |
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Kwiatowski pmid=7109038 |
| − | # | + | #Hallenbeck pmid=3546309 |
| − | # | + | #Petter pmid=8331067 |
| − | # | + | #Stummeyer pmid=15608653 |
| − | # | + | #Morely pmid=19411257 |
| + | #Steinbacher pmid=8023158 | ||
</biblio> | </biblio> | ||
| − | + | ||
[[Category:Glycoside Hydrolase Families|GH058]] | [[Category:Glycoside Hydrolase Families|GH058]] | ||
Latest revision as of 13:19, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH58 | |
| Clan | none |
| Mechanism | inverting |
| Active site residues | Arg596, Arg647 and Glu581 |
| CAZy DB link | |
| https://www.cazy.org/GH58.html | |
Substrate specificities
Glycoside hydrolases of family 58 are endo-N-acetylneuraminidases (also termed endo-sialidases). The capsular coat surrounding E. coli K1, is poly-α-2,8-sialic acid and protects the bacterium from the host immune system, but it also acts as an anchor point for bacteriophage infection. A range of bacteriophages specific to this E. coli capsule type have been described which have tailspike enzymes that degrade the capuslar material [1, 2, 3]
Kinetics and Mechanism
Family 58 endo-sialidases are specific for polysialic acid and form a unique family of endo-acting sialidases, all others previously reported being exo-acting. The recently determined structure of an endo-sialidase derived from bacteriophage K1F (endoNF) [4] revealed the active site to lack a number of the residues that are conserved in other sialidases, implying a new, endo-sialidase-specific catalytic mechanism. Using synthetic trifluoromethylumbelliferyl oligosialoside substrates kinetic parameters for hydrolysis at a single cleavage site were determined. Measurement of kcat/Km at a series of pH values revealed a dependence on a single protonated group of pKa 5. Direct 1H-NMR analysis of the hydrolysis of trifluoromethylumbelliferyl sialotrioside revealed that endoNF affects catalysis with an inverting mechanism [5].
Catalytic Residues
From the crystal structure of the GH-58 enzyme it was noted that the normal catalyic residues found in exo-sialidases were not present. Instead, three conserved amino acids (Arg596, Arg647 and Glu581) were observed that contribute to endosialidase activity. Single mutations of those residues reduced the level of activity, but not enough to mark the residues as absolutely required. However, when double mutants were examined there was a complete loss of hydrolysis, but binding to PSA still occured [4]. Since this enzyme proceeds with inverting stereochemistry, a catalytic base is implied by the mechanism, and Glu581 might fill that role [5].
Three-dimensional structures
The structure for the K1F tailspike enzyme has been solved by Stummeyer et al. [4]. The protein structure is similar to other phage tailspike endo-hydrolases in that it is an extended, trimeric molecule. The first tailspike enzyme to be studied in detail was the endo-rhamnosidase from the Salmonella phage P22 [6], which is in family GH90. The pictures here were generated in Pymol from PDB entry 1voe. Each monomer is a different colour to highlight the trimeric nature of the protein.
- PDB ID 1voe from GH58 (click images for large versions)
Family Firsts
- First sterochemistry determination
- The stereochemical outcome of this reaction was determined in the Withers laboratory at UBC using synthetic substrates with a trifluoromethyl-umbelliferol fluorophore [5]. This determination showed that unlike all other known sialidases, the endo-sialidase uses an inverting mechanism.
- First catalytic nucleophile identification
- Since this enzyme proceeds with inversion of sterechemistry, is does not use a catalytic nuceleophile.
- First general acid/base residue identification
- It has been suggested though not conclusively proven that Glu581 is the catalytic base for this reaction [4, 5].
- First 3-D structure
- The first structure determination was performed with a truncated version of the K1F protein [4].
References
- Kwiatkowski B, Boschek B, Thiele H, and Stirm S. (1982). Endo-N-acetylneuraminidase associated with bacteriophage particles. J Virol. 1982;43(2):697-704. DOI:10.1128/JVI.43.2.697-704.1982 |
- Hallenbeck PC, Vimr ER, Yu F, Bassler B, and Troy FA. (1987). Purification and properties of a bacteriophage-induced endo-N-acetylneuraminidase specific for poly-alpha-2,8-sialosyl carbohydrate units. J Biol Chem. 1987;262(8):3553-61. | Google Books | Open Library
- Petter JG and Vimr ER. (1993). Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J Bacteriol. 1993;175(14):4354-63. DOI:10.1128/jb.175.14.4354-4363.1993 |
- Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, and Ficner R. (2005). Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol. 2005;12(1):90-6. DOI:10.1038/nsmb874 |
- Morley TJ, Willis LM, Whitfield C, Wakarchuk WW, and Withers SG. (2009). A new sialidase mechanism: bacteriophage K1F endo-sialidase is an inverting glycosidase. J Biol Chem. 2009;284(26):17404-10. DOI:10.1074/jbc.M109.003970 |
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, and Reinemer P. (1994). Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science. 1994;265(5170):383-6. DOI:10.1126/science.8023158 |