CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Polysaccharide Lyase Family 2"
Wade Abbott (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (32 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Wade Abbott|Wade Abbott]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Wade Abbott|Wade Abbott]] |
---- | ---- | ||
| Line 11: | Line 11: | ||
|- | |- | ||
|{{Hl2}} colspan="2" align="center" |'''Polysaccharide Lyase Family PL2''' | |{{Hl2}} colspan="2" align="center" |'''Polysaccharide Lyase Family PL2''' | ||
| + | |- | ||
| + | |'''3D Structure''' | ||
| + | |(α/α)<sub>7</sub> barrel | ||
|- | |- | ||
|'''Mechanism''' | |'''Mechanism''' | ||
|β-elimination | |β-elimination | ||
|- | |- | ||
| − | |''' | + | |'''Charge neutraliser''' |
| − | | | + | |manganese |
|- | |- | ||
|'''Active site residues''' | |'''Active site residues''' | ||
| Line 30: | Line 33: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | PL2 activity has been demonstrated on | + | PL2 activity has been demonstrated on α-(1,4)-linked polygalacturonic acid (i.e. homogalacturonan or pectate) and α-(1,4)-linked oligogalacturonides <cite>Abbott2007, Shevchik1999</cite>. There are two subfamilies in PL2 <cite>#Lombard2010</cite>. Subfamily 1 is correlated with endolytic activity, whereas subfamily 2 is correlated with exolytic activity. Intriguingly, the majority of sequence entries are from the genomes of phytopathogenic or enteropathogenic bacteria, and are found in paralogous copies within each species <cite>Abbott2013</cite>. Several outliers exist, including the single copy PaePL2 from ''Paenibacillus sp.''Y412MC10, which may reflect the ancestral endolytic activity <cite>Abbott2013</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | + | Use of a β-elimination reaction to cleave the glycosidic bonds in pectate requires a Brønstead base for proton abstraction and a catalytic metal (e.g. Mn<sup>2+</sup> or Mg<sup>2+</sup>) for acidification of the β-proton and oxyanion stabilization. PL2s have reported pH optimas in the range of 7.4 - 9.6 <cite>Abbott2007, Abbott2013</cite>, which is substantially lower than the p''K''<sub>a</sub> of arginine. These effects have been attributed to localized p''K''<sub>a</sub> effects within the active site. β-elimination results in the production of a new reducing end (residue in the -1 subsite) and a 4,5-unsaturated bond in the other nascent sugar chain end (residue in the +1 subsite). Full kinetics with a library of metals have been performed for the YePL2A and YePL2B <cite>McLean2015</cite>. Both paralogs have the best catalytic efficiency with Mn<sup>2+</sup>; however, the secreted YePL2A demonstrates more plasticity in metal utilization; whereas, the cytoplasmic YePL2B is selective for Mn<sup>2+</sup>. | |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | The Brønstead base for the PL2 family is an | + | The Brønstead base for the PL2 family is an arginine, which is consistent with most pectate lyase families. R171 in YePL2A was the first catalytic base described for the family and it is completely conserved within the family <cite>Abbott2007, Abbott2013</cite>. The metal coordination pocket in YePL2A consists of two histidine residues (YePL2A: H109 and H172) and one glutamic acid (YePL2A: E130). |
| − | |||
| − | |||
| − | |||
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The structure of the endolytic PL2A from ''Yersinia enterocolitica'' (YePL2A) | + | [[Image:PL2A.png|thumb|350px|[{{PDBlink}}2v8j YePL2A] in complex with Mn<sup>2+</sup>]]The structure of the endolytic PL2A from ''Yersinia enterocolitica'' (YePL2A) was the first PL2 structure to be reported <cite>Abbott2007</cite>. In this study, structural differences were noted between a native-form ([{{PDBlink}}2v8i PDB 2v8i], 1.50 Å), and complexes with trigalacturonate ([{{PDBlink}}2v8k PDB 2v8k], 2.1 Å) and a transition metal ([{{PDBlink}}2v8j PDB 2v8j], 2.01 Å). Family 2 PLs adopt a rare α/α<sub>7</sub> barrel fold, with an active site cleft extending along the surface of the enzyme between two catalytic arms. Substrate binding induces a conformational change and the arms close about the substrate. |
== Family Firsts == | == Family Firsts == | ||
| − | ;First catalytic activity: PelY from ''Yersinia pseudotuberculosis'' macerated cucumber <cite>Manulis1988</cite>. | + | ;First catalytic activity: PelY/YpsPL2 from ''Yersinia pseudotuberculosis'' macerated cucumber <cite>Manulis1988</cite>. |
| − | ;First catalytic base identification: YePL2A (YE4069) | + | ;First catalytic base identification: YePL2A (YE4069) R171 from ''Yersinia enterocolitica'' <cite>Abbott2007</cite>. |
| − | ;First catalytic divalent cation identification: DdPL2 | + | ;First catalytic divalent cation identification: PelW/DdPL2 (Dda3937_03361) from ''Dickeya Dadantii'' 3937 (previously ''Erwinia chrysanthemi''3937) <cite>Shevchik1999</cite>. |
| − | ;First 3-D structure: | + | ;First 3-D structure: YePL2A (YE4069) from ''Yersinia enterocolitica'' <cite>Abbott2007</cite> ([{{PDBlink}}2v8i PDB 2v8i], [{{PDBlink}}2v8j PDB 2v8j], [{{PDBlink}}2v8k PDB 2v8k]). |
== References == | == References == | ||
| Line 57: | Line 57: | ||
#Manulis1988 pmid=2832382 | #Manulis1988 pmid=2832382 | ||
#Abbott2013 pmid=24013861 | #Abbott2013 pmid=24013861 | ||
| + | #Lombard2010 pmid=20925655 | ||
| + | #McLean2015 pmid=26160170 | ||
</biblio> | </biblio> | ||
[[Category:Polysaccharide Lyase Families|PL002]] | [[Category:Polysaccharide Lyase Families|PL002]] | ||
Latest revision as of 13:17, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Polysaccharide Lyase Family PL2 | |
| 3D Structure | (α/α)7 barrel |
| Mechanism | β-elimination |
| Charge neutraliser | manganese |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/PL2.html | |
Substrate specificities
PL2 activity has been demonstrated on α-(1,4)-linked polygalacturonic acid (i.e. homogalacturonan or pectate) and α-(1,4)-linked oligogalacturonides [1, 2]. There are two subfamilies in PL2 [3]. Subfamily 1 is correlated with endolytic activity, whereas subfamily 2 is correlated with exolytic activity. Intriguingly, the majority of sequence entries are from the genomes of phytopathogenic or enteropathogenic bacteria, and are found in paralogous copies within each species [4]. Several outliers exist, including the single copy PaePL2 from Paenibacillus sp.Y412MC10, which may reflect the ancestral endolytic activity [4].
Kinetics and Mechanism
Use of a β-elimination reaction to cleave the glycosidic bonds in pectate requires a Brønstead base for proton abstraction and a catalytic metal (e.g. Mn2+ or Mg2+) for acidification of the β-proton and oxyanion stabilization. PL2s have reported pH optimas in the range of 7.4 - 9.6 [1, 4], which is substantially lower than the pKa of arginine. These effects have been attributed to localized pKa effects within the active site. β-elimination results in the production of a new reducing end (residue in the -1 subsite) and a 4,5-unsaturated bond in the other nascent sugar chain end (residue in the +1 subsite). Full kinetics with a library of metals have been performed for the YePL2A and YePL2B [5]. Both paralogs have the best catalytic efficiency with Mn2+; however, the secreted YePL2A demonstrates more plasticity in metal utilization; whereas, the cytoplasmic YePL2B is selective for Mn2+.
Catalytic Residues
The Brønstead base for the PL2 family is an arginine, which is consistent with most pectate lyase families. R171 in YePL2A was the first catalytic base described for the family and it is completely conserved within the family [1, 4]. The metal coordination pocket in YePL2A consists of two histidine residues (YePL2A: H109 and H172) and one glutamic acid (YePL2A: E130).
Three-dimensional structures
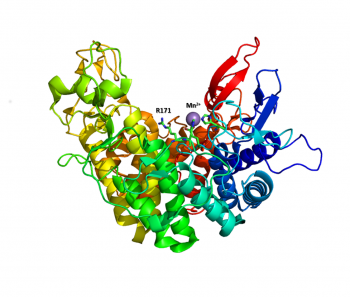
The structure of the endolytic PL2A from Yersinia enterocolitica (YePL2A) was the first PL2 structure to be reported [1]. In this study, structural differences were noted between a native-form (PDB 2v8i, 1.50 Å), and complexes with trigalacturonate (PDB 2v8k, 2.1 Å) and a transition metal (PDB 2v8j, 2.01 Å). Family 2 PLs adopt a rare α/α7 barrel fold, with an active site cleft extending along the surface of the enzyme between two catalytic arms. Substrate binding induces a conformational change and the arms close about the substrate.
Family Firsts
- First catalytic activity
- PelY/YpsPL2 from Yersinia pseudotuberculosis macerated cucumber [6].
- First catalytic base identification
- YePL2A (YE4069) R171 from Yersinia enterocolitica [1].
- First catalytic divalent cation identification
- PelW/DdPL2 (Dda3937_03361) from Dickeya Dadantii 3937 (previously Erwinia chrysanthemi3937) [2].
- First 3-D structure
- YePL2A (YE4069) from Yersinia enterocolitica [1] (PDB 2v8i, PDB 2v8j, PDB 2v8k).
References
- Abbott DW and Boraston AB. (2007). A family 2 pectate lyase displays a rare fold and transition metal-assisted beta-elimination. J Biol Chem. 2007;282(48):35328-36. DOI:10.1074/jbc.M705511200 |
- Shevchik VE, Condemine G, Robert-Baudouy J, and Hugouvieux-Cotte-Pattat N. (1999). The exopolygalacturonate lyase PelW and the oligogalacturonate lyase Ogl, two cytoplasmic enzymes of pectin catabolism in Erwinia chrysanthemi 3937. J Bacteriol. 1999;181(13):3912-9. DOI:10.1128/JB.181.13.3912-3919.1999 |
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, and Henrissat B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432(3):437-44. DOI:10.1042/BJ20101185 |
- Abbott DW, Thomas D, Pluvinage B, and Boraston AB. (2013). An ancestral member of the polysaccharide lyase family 2 displays endolytic activity and magnesium dependence. Appl Biochem Biotechnol. 2013;171(7):1911-23. DOI:10.1007/s12010-013-0483-9 |
- McLean R, Hobbs JK, Suits MD, Tuomivaara ST, Jones DR, Boraston AB, and Abbott DW. (2015). Functional Analyses of Resurrected and Contemporary Enzymes Illuminate an Evolutionary Path for the Emergence of Exolysis in Polysaccharide Lyase Family 2. J Biol Chem. 2015;290(35):21231-43. DOI:10.1074/jbc.M115.664847 |
- Manulis S, Kobayashi DY, and Keen NT. (1988). Molecular cloning and sequencing of a pectate lyase gene from Yersinia pseudotuberculosis. J Bacteriol. 1988;170(4):1825-30. DOI:10.1128/jb.170.4.1825-1830.1988 |