CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 3"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) |
||
| Line 30: | Line 30: | ||
The GH3 [[glycoside hydrolase]] family currently groups together exo-acting β-D-glucosidases, α-L-arabinofuranosidases, β-D-xylopyranosidases, ''N''-acetyl-β-D-glucosaminidases, and ''N''-acetyl-β-D-glucosaminide phosphorylases <cite>Harvey2000 Macdonald2015</cite>. Widely distributed in bacteria, fungi and plants, GH3 enzymes carry out a range of functions including cellulosic biomass degradation, plant and bacterial cell wall remodeling, energy metabolism and pathogen defense. In many cases the enzymes have dual or broad substrate specificities with respect to monosaccharide residues, linkage position and chain length of the substrate. For example, there are several well-characterized ‘bifunctional’ enzymes in the family that have both α-L-arabinofuranosidase and β-D-xylopyranosidase activity <cite>Lee2003</cite>, and one characterized example of an ''N''-acetyl-β-D-glucosaminide/β-glucoside [[glycoside hydrolase|hydrolase]]/[[phosphorylases|phosphorylase]] from ''Cellulomonas fimi'' (Nag3) <cite>Mayer2006 Macdonald2015</cite>. GH3 β-D-glucosidases from barley, which are more precisely referred to as β-D-glucan glucohydrolases, are also broad specificity exo-hydrolases that remove single glucosyl residues from the non-reducing ends of a range of β-D-glucans, β-D-oligoglucosides and aryl β-D-glucosides, including (1,3)-β-D-glucans, (1,4)-β-D-glucans, (1,3;1,4)-β-D-glucans and (1,6)-β-D-glucans, 4-nitrophenyl β-D-glucoside, certain cyanogenic β-D-glucosides and some β-D-oligoxyloglucosides <cite>Hrmova1998</cite>. | The GH3 [[glycoside hydrolase]] family currently groups together exo-acting β-D-glucosidases, α-L-arabinofuranosidases, β-D-xylopyranosidases, ''N''-acetyl-β-D-glucosaminidases, and ''N''-acetyl-β-D-glucosaminide phosphorylases <cite>Harvey2000 Macdonald2015</cite>. Widely distributed in bacteria, fungi and plants, GH3 enzymes carry out a range of functions including cellulosic biomass degradation, plant and bacterial cell wall remodeling, energy metabolism and pathogen defense. In many cases the enzymes have dual or broad substrate specificities with respect to monosaccharide residues, linkage position and chain length of the substrate. For example, there are several well-characterized ‘bifunctional’ enzymes in the family that have both α-L-arabinofuranosidase and β-D-xylopyranosidase activity <cite>Lee2003</cite>, and one characterized example of an ''N''-acetyl-β-D-glucosaminide/β-glucoside [[glycoside hydrolase|hydrolase]]/[[phosphorylases|phosphorylase]] from ''Cellulomonas fimi'' (Nag3) <cite>Mayer2006 Macdonald2015</cite>. GH3 β-D-glucosidases from barley, which are more precisely referred to as β-D-glucan glucohydrolases, are also broad specificity exo-hydrolases that remove single glucosyl residues from the non-reducing ends of a range of β-D-glucans, β-D-oligoglucosides and aryl β-D-glucosides, including (1,3)-β-D-glucans, (1,4)-β-D-glucans, (1,3;1,4)-β-D-glucans and (1,6)-β-D-glucans, 4-nitrophenyl β-D-glucoside, certain cyanogenic β-D-glucosides and some β-D-oligoxyloglucosides <cite>Hrmova1998</cite>. | ||
| − | In contrast to the broad substrate specificities observed for the GH3 enzymes described above, GH3 ''N''-acetyl-β-D-glucosaminidases are selective for ''N''-acetyl-β-D-glucosamine (GlcNAc) <cite>Chitlaru1996</cite> (though exceptions exist, e.g. ''Cellulomonas fimi'' Nag3 <cite>Mayer2006</cite>). A notable GH3 ''N''-acetyl-β-D-glucosaminidase of prokaryotes is NagZ, which participates in bacterial cell wall recycling (for review see <cite>Johnson2013</cite>) by removing GlcNAc from 1,6-anhydroMurNAc-peptides in Gram-negative bacteria <cite>Cheng2000</cite>, or GlcNAc from GlcNAc-MurNAc-peptides in Gram-positive bacteria <cite>Litzinger2010b</cite>. The NagZ product 1,6-anhydroMurNAc-peptide is an important activator of AmpC β-lactamase hyper-production in several Gram-negative pathogens, making the enzyme of interest as a potential therapeutic target <cite>Mark2011</cite>. | + | In contrast to the broad substrate specificities observed for the GH3 enzymes described above, GH3 ''N''-acetyl-β-D-glucosaminidases are selective for ''N''-acetyl-β-D-glucosamine (GlcNAc) <cite>Chitlaru1996 Ducatti2016</cite> (though exceptions exist, e.g. ''Cellulomonas fimi'' Nag3 <cite>Mayer2006</cite>). A notable GH3 ''N''-acetyl-β-D-glucosaminidase of prokaryotes is NagZ, which participates in bacterial cell wall recycling (for review see <cite>Johnson2013</cite>) by removing GlcNAc from 1,6-anhydroMurNAc-peptides in Gram-negative bacteria <cite>Cheng2000</cite>, or GlcNAc from GlcNAc-MurNAc-peptides in Gram-positive bacteria <cite>Litzinger2010b</cite>. The NagZ product 1,6-anhydroMurNAc-peptide is an important activator of AmpC β-lactamase hyper-production in several Gram-negative pathogens, making the enzyme of interest as a potential therapeutic target <cite>Mark2011</cite>. |
Due to the high diversity of protein structural arrangements found among GH3 members (see below), the phylogeny of this family is complex. Classification of GH3 members into subfamilies has been performed previously <cite>Harvey2000 Cournoyer2003</cite>, however a robust subfamily classification (on par with those for [[GH13]] <cite>Stam2006</cite>, [[GH5]] <cite>Aspeborg2012</cite>, and the polysaccharide lyases <cite>Lombard2010</cite>), is currently not available. However, as more plant genome sequences are published, it is becoming clear that the GH3 glycoside hydrolases in plants are encoded by multi-gene families. In both monocot and dicot species, there are 15-18 genes for the GH3 enzymes, while in lower plants such as the Physcomitrella and Selaginella mosses 6-7 genes encoding GH3 glycoside hydrolases have been identified (<cite>Henrissat2001 GeislerLee2006</cite>, see also [http://www.phytozome.org/ Phytozome] <cite>Goodstein2012</cite>). Plant β-''N''-acetylglucosaminidases have not been identified in GH3 thus far. | Due to the high diversity of protein structural arrangements found among GH3 members (see below), the phylogeny of this family is complex. Classification of GH3 members into subfamilies has been performed previously <cite>Harvey2000 Cournoyer2003</cite>, however a robust subfamily classification (on par with those for [[GH13]] <cite>Stam2006</cite>, [[GH5]] <cite>Aspeborg2012</cite>, and the polysaccharide lyases <cite>Lombard2010</cite>), is currently not available. However, as more plant genome sequences are published, it is becoming clear that the GH3 glycoside hydrolases in plants are encoded by multi-gene families. In both monocot and dicot species, there are 15-18 genes for the GH3 enzymes, while in lower plants such as the Physcomitrella and Selaginella mosses 6-7 genes encoding GH3 glycoside hydrolases have been identified (<cite>Henrissat2001 GeislerLee2006</cite>, see also [http://www.phytozome.org/ Phytozome] <cite>Goodstein2012</cite>). Plant β-''N''-acetylglucosaminidases have not been identified in GH3 thus far. | ||
Revision as of 08:26, 6 March 2017
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Authors: ^^^Geoff Fincher^^^, ^^^Brian Mark^^^, and ^^^Harry Brumer^^^
- Responsible Curator: ^^^Bernard Henrissat^^^
| Glycoside Hydrolase Family GH3 | |
| Clan | none |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH3.html | |
Substrate specificities
The GH3 glycoside hydrolase family currently groups together exo-acting β-D-glucosidases, α-L-arabinofuranosidases, β-D-xylopyranosidases, N-acetyl-β-D-glucosaminidases, and N-acetyl-β-D-glucosaminide phosphorylases [1, 2]. Widely distributed in bacteria, fungi and plants, GH3 enzymes carry out a range of functions including cellulosic biomass degradation, plant and bacterial cell wall remodeling, energy metabolism and pathogen defense. In many cases the enzymes have dual or broad substrate specificities with respect to monosaccharide residues, linkage position and chain length of the substrate. For example, there are several well-characterized ‘bifunctional’ enzymes in the family that have both α-L-arabinofuranosidase and β-D-xylopyranosidase activity [3], and one characterized example of an N-acetyl-β-D-glucosaminide/β-glucoside hydrolase/phosphorylase from Cellulomonas fimi (Nag3) [2, 4]. GH3 β-D-glucosidases from barley, which are more precisely referred to as β-D-glucan glucohydrolases, are also broad specificity exo-hydrolases that remove single glucosyl residues from the non-reducing ends of a range of β-D-glucans, β-D-oligoglucosides and aryl β-D-glucosides, including (1,3)-β-D-glucans, (1,4)-β-D-glucans, (1,3;1,4)-β-D-glucans and (1,6)-β-D-glucans, 4-nitrophenyl β-D-glucoside, certain cyanogenic β-D-glucosides and some β-D-oligoxyloglucosides [5].
In contrast to the broad substrate specificities observed for the GH3 enzymes described above, GH3 N-acetyl-β-D-glucosaminidases are selective for N-acetyl-β-D-glucosamine (GlcNAc) [6, 7] (though exceptions exist, e.g. Cellulomonas fimi Nag3 [4]). A notable GH3 N-acetyl-β-D-glucosaminidase of prokaryotes is NagZ, which participates in bacterial cell wall recycling (for review see [8]) by removing GlcNAc from 1,6-anhydroMurNAc-peptides in Gram-negative bacteria [9], or GlcNAc from GlcNAc-MurNAc-peptides in Gram-positive bacteria [10]. The NagZ product 1,6-anhydroMurNAc-peptide is an important activator of AmpC β-lactamase hyper-production in several Gram-negative pathogens, making the enzyme of interest as a potential therapeutic target [11].
Due to the high diversity of protein structural arrangements found among GH3 members (see below), the phylogeny of this family is complex. Classification of GH3 members into subfamilies has been performed previously [1, 12], however a robust subfamily classification (on par with those for GH13 [13], GH5 [14], and the polysaccharide lyases [15]), is currently not available. However, as more plant genome sequences are published, it is becoming clear that the GH3 glycoside hydrolases in plants are encoded by multi-gene families. In both monocot and dicot species, there are 15-18 genes for the GH3 enzymes, while in lower plants such as the Physcomitrella and Selaginella mosses 6-7 genes encoding GH3 glycoside hydrolases have been identified ([16, 17], see also Phytozome [18]). Plant β-N-acetylglucosaminidases have not been identified in GH3 thus far.
Kinetics and Mechanism
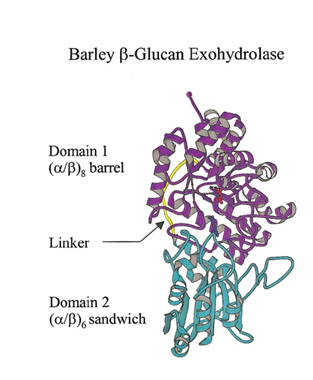
GH3 glycoside hydrolases remove single glycosyl residues from the non-reducing ends of their substrates. Catalysis occurs via a classical Koshland double-displacement mechanism with the anomeric configuration of the released glycose being retained. The retention of anomeric configuration has been established experimentally for several enzymes (see [2, 7] and references therein). The active site of GH3 enzymes consists of two glucosyl-binding subsites (-1 and +1) with an enzymic nucleophile and general acid/base residue flanking the junction of these two subsites (see [19] for subsite nomenclature). This arrangement of catalytic residues was first visualized in 1999 when the crystal structure of barley β-D-glucan glucohydrolase was determined in complex with glucose [20] (Fig. 1).
Several studies have contributed to the understanding of the kinetics and mechanism of GH3 enzymes, including detailed analyses of β-glucosidases from fungi (Aspergillus wentii [21] and Aspergillus niger [22, 23]) and Gram-negative bacteria (Flavobacterium meningosepticum [24, 25], Thermotoga neapolitana [26]), as well as a GH3 glucosylceramidase from the Gram-positive microbe Paenibacillus sp. TS12 [27]. Kinetic and mechanistic analyses of β-D-glucan glucohydrolases and two ‘bifunctional’ α-L-arabinofuranosidase/β-D-xylopyranosidases from plants (barley) have also been carried out [3, 5]. These studies, combined with kinetic and mechanistic analyses of N-acetyl-β-D-glucosaminidases from the Gram-positive microbe Bacilus subtillus [10, 28], and Gram-negative microbes Vibrio furnisii [29, 30], Vibrio cholerea [31] and Salmonella typhimerium [28] confirm that while the catalytic nucleophile of GH3 enzymes is well conserved, the location and identity of the general acid/base residue is not (see below).
Kinetic partitioning of the glycosyl-enzyme intermediate between hydrolysis and transglycosylation has been studied in detail for a number of fungal GH3 enzymes [32]. Notably, the N-acetylglucosaminidase Nag3 from Cellulomonas fimi may use either water (functioning as a hydrolase) or phosphate (functioning as a phosphorylase) as an acceptor in the breakdown of GlcNAc-enzyme and Glc-enzyme intermediates [2] (see also Catalytic acid/base section, below).
Catalytic Residues
Catalytic nucleophile
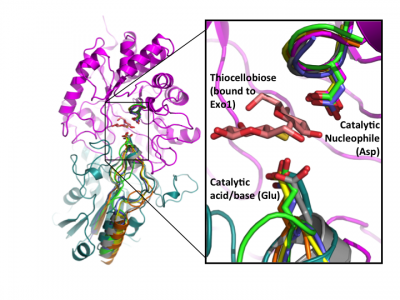
Early labeling experiments of a β-glucosidase from Aspergillus wentii using conduritol B-epoxide by Bause and Legler in 1974 [21] suggested an aspartate residue within the sequence VMSDW as the putative catalytic nucleophile long before the establishment of the CAZy classification [34] and recognition of this enzyme as a GH3 member. The homologous residue was later implicated by the crystal structure of barley β-D-glucan glucohydrolase, which identified Asp285 within the sequence GFVISDW as being appropriately positioned to act as a nucleophile during catalysis [20]. Direct identification of this aspartate as the catalytic nucleophile via active-site labeling using 2-deoxy-2-fluoro-β-D-glycosides of GH3 β-glucosidases from A. niger [22] and F. meningosepticum [24], a glucosylceramidase from Paenibacillus sp. [27], as well as the barley β-D-glucan glucohydrolase (including conduritol B-epoxide labelling and crystallography of both inactive complexes) [35], and labeling of NagZ enzymes from V. furnisii [29] and S. typhimerium [28] using 2-acetamido-2-deoxy-5-fluoro-β-D-glucopyranosyl fluoride, have confirmed that this catalytic residue is well conserved in GH3 enzymes from diverse species. The conservation of this aspartate becomes clear when known crystal structures of a number of GH3 enzymes are superposed (Fig. 2).
Catalytic acid/base
Though sequence alignments can now be used to easily identify the catalytic nucleophile of GH3 members, the position of the general acid/base residue is phylogenetically variable and less readily identifiable [23]. Detailed kinetic analyses and an increase in 3D structural information continue to provide insight into identity of the elusive general acid-base residue across the diversity of GH3 members.
The seminal structural study of the barley β-D-glucan glucohydrolase ExoI, in complex with the product glucose, was the first to suggest the identity of the catalytic acid/base in a GH3 member. Barley ExoI is a two-domain enzyme consisting of an N-terminal (β/α)8 barrel domain housing the active site pocket and the catalytic nucleophile (Asp285). The C-terminal domain contains a glutamate residue (Glu 491) projecting into the active site of the (β/α)8 barrel domain, which was proposed to act as the catalytic acid/base [20]. These assignments were also supported by crystallography of pseudo-Michaelis complexes with non-hydrolyzable thio-glycosides [35, 36].
Kinetic analyses, including the use of site-directed mutation and chemical rescue, have been used to provide early experimental support for the identification of the catalytic acid/base in several bacterial and fungal GH3 members: Flavobacterium meningosepticum β-glucosidase [25], Paenibacillus sp. TS12 glucosylceramidase [27], Thermotoga neapolitana β-glucosidase [26], and Aspergillus niger β-glucosidase [23]. The identification of Glu-473 as the catalytic acid/base in the F. meningosepticum enzyme was further supported by covalent labelling with N-bromoacetyl-β-D-glucosylamine and peptide mass spectrometry [24].
Later structural studies of bacterial and fungal GH3 enzymes have since shown that while insertion of additional domains is possible (which shifts the location of the glutamic acid general acid-base within the primary sequence), the contribution of two-domains to the active site architecture, as first observed for the barley enzyme, appears to be a core feature of multidomain GH3 β-glucanases [26, 37, 38]. Superposition of available GH3 β-D-glucan glucohydrolase structures clearly reveals the conservation of this architecture, and directly highlights active-site residue homology in the absence of protein sequence similarity - especially so in the case of the catalytic acid-base [23] (Fig. 2).
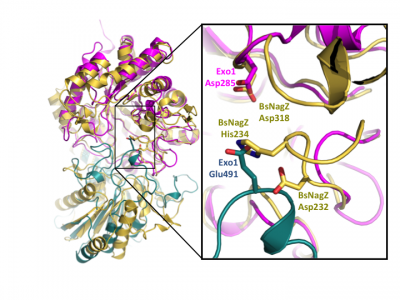
Notably, GH3 NagZ enzymes represent a significant departure from the above two-domain active site architecture paradigm. A crystal structure of NagZ from B. subtilis, together with kinetic analysis, provided evidence that the catalytic acid-base is an unusual histidine/aspartate dyad that resides within a flexible loop on the catalytic (β/α)8 barrel, not on a separate domain [33]. Though the enzyme adopts a two-domain fold similar to the barely β-D-glucan glucohydrolase, the C-terminal domain does not participate in catalysis (Fig. 3). In fact, most NagZ enzymes from Gram-negative bacteria are single domain enzymes comprised solely of a catalytic (β/α)8 barrel that contains the conserved aspartate nucleophile and a catalytic histidine/aspartate dyad on flexible loop, as seen for B. subtilis NagZ [28] (Fig. 4). The flexible loop containing the His/Asp dyad is unique to GH3 NagZ enzymes and can be identified by the consensus motif [KH(F/I)PG(H/L)GXXXXD(S/T)H] (catalytic dyad highlighted in boldface) [29]. In light of these studies, the residue identification in a Clostridium paraputrificum N-acetyl-β-D-glucosaminidase (Nag3A) would appear to be unreliable [39].
In 2015, a study revisiting the mechanism of Cellulomonas fimi N-acetylglucosaminidase Nag3, which belongs to the same subfamily as the aforementioned enzymes and likewise has the conserved His/Asp dyad, indicated that Nag3 is predominantly a glycoside phosphorylase, rather than a glycoside hydrolase [2]. From this study, it was proposed that all members of the NagZ subfamily are phosphorylases, and that the catalytic histidine is employed to avoid Coulombic repulsion with the incoming acceptor substrate, viz. phosphate [2]. However, a more recent study has indicated that not all members of this subfamily are phosphorylases, which casts doubt on the generality of this proposal: The presence of phosphate does not alter the kinetics of a Herbaspirillum seropedicae SmR1 beta-N-acetylglucosaminidase, and only the hydrolysis product is observed [7].
Three-dimensional structures
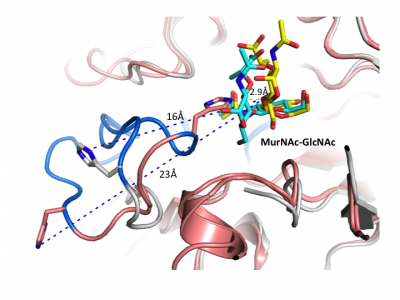
Crystal structures are now available for a number of multidomain GH3 members, including barley β-D-glucan glucohydrolase [20] and a mechanistically related β-glucanase from the marine bacterium Pseudoalteromonas sp. [37], as well as β-glucosidases from Kluyveromyces marxianus (KmBglI) [38] , Trichoderma reesei (Cel3A) (PDB 4I8D (unpublished)), Thermotoga neapolitana [26] and a macrolide β-glycosidase / β-glucosidase (DesR) from Streptomyces venezuelae [40] (please see the GH3 structure page of the CAZy DB for a continuously updated list). These structures have revealed considerable diversity in architecture and arrangement of domains in GH3 enzymes that are present in addition to the core catalytic domain, and have provided insight into how these domains affect function.
Crystal structures of a number of NagZ enzymes are also available, including the two-domain NagZ from B. subtilis [33] and single domain NagZ enzymes from Vibrio cholera (PDB 1TR9 (unpublished)) & [31], S. typhimurium [28], Deinococcus radiodurans (PDB 3TEV (unpublished)) and Burkholderia cenocepacia (PDB 4GNV (unpublished)). Given the role of NagZ in the activation of AmpC-mediated β-lactam resistance in Gram-negative bacteria [11], a number of crystallographic and kinetic studies have focused on the development of small-molecule inhibitors of NagZ [31, 41, 42], some of which have been designed to be selective for GH3 NagZ over functionally related human enzymes from families GH20 and GH84 [31, 41].
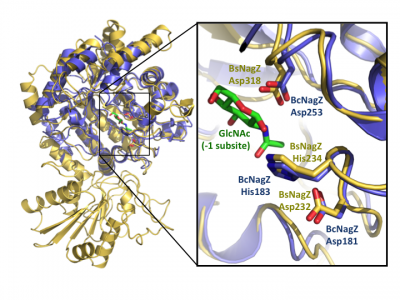
Insight into the broad specificity observed for some GH3 enzymes has been provided from X-ray crystallographic data and from molecular modeling of enzyme-substrate complexes of the barley β-D-glucan glucohydrolase [20, 36]. Recent structural studies of NagZ enzymes from S. typhimurium and B. subtilis have shown the flexible catalytic loop participates in distorting the bound terminal GlcNAc sugar toward a 1S3 conformation during catalysis [28] (Fig. 5). Distortion of the substrate toward a 1S3 conformation is in agreement with other retaining β-glycosidases; however, this has not been observed for other GH3 enzymes, in which the substrate has been found in a relaxed, near-4C1 conformation.
Family Firsts
- First sterochemistry determination
- To be added.
- First catalytic nucleophile identification
- First suggested by Bause and Legler in 1974 using conduritol B-epoxide labelling of an Aspergillus wentii glucosidase [21], and later supported by the crystal structures of a product complex of a barley β-D-glucan glucohydrolase [20] and a trapped covalent intermediate [35]. Contemporaneous active-site labeling of an A. niger β-glucosidase [22] and V. furnisii NagZ [29] using 2-deoxy-2-fluoro-β-D-glycosides allowed unequivocal identification of the catalytic nucleophiles in these enzymes.
- First general acid/base identification
- In contrast to the catalytic nucleophile, the catalytic acid/base is not broadly conserved on the protein sequence level; thus, the "first" identification was not strictly definitive in this family (this living history is detailed above). The GH3 general acid/base was first suggested by a product complex of a barley β-D-glucan glucohydrolase [20] (and later by other complex structures of this same enzyme [35, 36]). The earliest definitive kinetic studies were performed on a Flavobacterium meningosepticum β-glucosidase [24, 25]. The first revelation of the atypical Asp/His dyad fulfilling this role was for B. subtilis NagZ [10].
- First 3-D structure
- The first 3D structure in family GH3 was that of the two-domain barley β-D-glucan glucohydrolase [20]. As discussed above, the number and organization of domains among GH3 members is diverse, such that a number of seminal structures could be highlighted.
References
- Harvey AJ, Hrmova M, De Gori R, Varghese JN, and Fincher GB. (2000). Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins. 2000;41(2):257-69. DOI:10.1002/1097-0134(20001101)41:2<257::aid-prot100>3.0.co;2-c |
- Macdonald SS, Blaukopf M, and Withers SG. (2015). N-acetylglucosaminidases from CAZy family GH3 are really glycoside phosphorylases, thereby explaining their use of histidine as an acid/base catalyst in place of glutamic acid. J Biol Chem. 2015;290(8):4887-4895. DOI:10.1074/jbc.M114.621110 |
- Lee RC, Hrmova M, Burton RA, Lahnstein J, and Fincher GB. (2003). Bifunctional family 3 glycoside hydrolases from barley with alpha -L-arabinofuranosidase and beta -D-xylosidase activity. Characterization, primary structures, and COOH-terminal processing. J Biol Chem. 2003;278(7):5377-87. DOI:10.1074/jbc.M210627200 |
- Mayer C, Vocadlo DJ, Mah M, Rupitz K, Stoll D, Warren RA, and Withers SG. (2006). Characterization of a beta-N-acetylhexosaminidase and a beta-N-acetylglucosaminidase/beta-glucosidase from Cellulomonas fimi. FEBS J. 2006;273(13):2929-41. DOI:10.1111/j.1742-4658.2006.05308.x |
-
Hrmova, M. and Fincher, G.B. (1998) Barley ß-D-glucan exohydrolases. Substrate specificity and kinetic properties. Carbohydr. Res. 305, 209-221. DOI:10.1016/S0008-6215(97)00257-7
- Chitlaru E and Roseman S. (1996). Molecular cloning and characterization of a novel beta-N-acetyl-D-glucosaminidase from Vibrio furnissii. J Biol Chem. 1996;271(52):33433-9. DOI:10.1074/jbc.271.52.33433 |
- Ducatti DR, Carroll MA, and Jakeman DL. (2016). On the phosphorylase activity of GH3 enzymes: A β-N-acetylglucosaminidase from Herbaspirillum seropedicae SmR1 and a glucosidase from Saccharopolyspora erythraea. Carbohydr Res. 2016;435:106-112. DOI:10.1016/j.carres.2016.09.015 |
- Johnson JW, Fisher JF, and Mobashery S. (2013). Bacterial cell-wall recycling. Ann N Y Acad Sci. 2013;1277(1):54-75. DOI:10.1111/j.1749-6632.2012.06813.x |
- Cheng Q, Li H, Merdek K, and Park JT. (2000). Molecular characterization of the beta-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J Bacteriol. 2000;182(17):4836-40. DOI:10.1128/JB.182.17.4836-4840.2000 |
- Litzinger S, Duckworth A, Nitzsche K, Risinger C, Wittmann V, and Mayer C. (2010). Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by beta-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. J Bacteriol. 2010;192(12):3132-43. DOI:10.1128/JB.01256-09 |
- Mark BL, Vocadlo DJ, and Oliver A. (2011). Providing β-lactams a helping hand: targeting the AmpC β-lactamase induction pathway. Future Microbiol. 2011;6(12):1415-27. DOI:10.2217/fmb.11.128 |
- Cournoyer B and Faure D. (2003). Radiation and functional specialization of the family-3 glycoside hydrolases. J Mol Microbiol Biotechnol. 2003;5(3):190-8. DOI:10.1159/000070269 |
- Stam MR, Danchin EG, Rancurel C, Coutinho PM, and Henrissat B. (2006). Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng Des Sel. 2006;19(12):555-62. DOI:10.1093/protein/gzl044 |
- Aspeborg H, Coutinho PM, Wang Y, Brumer H 3rd, and Henrissat B. (2012). Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol. 2012;12:186. DOI:10.1186/1471-2148-12-186 |
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, and Henrissat B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432(3):437-44. DOI:10.1042/BJ20101185 |
- Henrissat B, Coutinho PM, and Davies GJ. (2001). A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol. 2001;47(1-2):55-72. | Google Books | Open Library
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunnerås S, Sundberg B, Karpinski S, Teeri TT, Kleczkowski LA, Henrissat B, and Mellerowicz EJ. (2006). Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 2006;140(3):946-62. DOI:10.1104/pp.105.072652 |
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, and Rokhsar DS. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178-86. DOI:10.1093/nar/gkr944 |
- Davies GJ, Wilson KS, and Henrissat B. (1997). Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321 ( Pt 2)(Pt 2):557-9. DOI:10.1042/bj3210557 |
- Varghese JN, Hrmova M, and Fincher GB. (1999). Three-dimensional structure of a barley beta-D-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure. 1999;7(2):179-90. DOI:10.1016/s0969-2126(99)80024-0 |
- Bause E and Legler G. (1974). Isolation and amino acid sequence of a hexadecapeptide from the active site of beta-glucosidase A3 from Aspergillus wentii. Hoppe Seylers Z Physiol Chem. 1974;355(4):438-42. DOI:10.1515/bchm2.1974.355.1.438 |
- Dan S, Marton I, Dekel M, Bravdo BA, He S, Withers SG, and Shoseyov O. (2000). Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger beta-glucosidase. J Biol Chem. 2000;275(7):4973-80. DOI:10.1074/jbc.275.7.4973 |
- Thongpoo P, McKee LS, Araújo AC, Kongsaeree PT, and Brumer H. (2013). Identification of the acid/base catalyst of a glycoside hydrolase family 3 (GH3) beta-glucosidase from Aspergillus niger ASKU28. Biochim Biophys Acta. 2013;1830(3):2739-49. DOI:10.1016/j.bbagen.2012.11.014 |
- Chir J, Withers S, Wan CF, and Li YK. (2002). Identification of the two essential groups in the family 3 beta-glucosidase from Flavobacterium meningosepticum by labelling and tandem mass spectrometric analysis. Biochem J. 2002;365(Pt 3):857-63. DOI:10.1042/BJ20020186 |
- Li YK, Chir J, Tanaka S, and Chen FY. (2002). Identification of the general acid/base catalyst of a family 3 beta-glucosidase from Flavobacterium meningosepticum. Biochemistry. 2002;41(8):2751-9. DOI:10.1021/bi016049e |
- Pozzo T, Pasten JL, Karlsson EN, and Logan DT. (2010). Structural and functional analyses of beta-glucosidase 3B from Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3. J Mol Biol. 2010;397(3):724-39. DOI:10.1016/j.jmb.2010.01.072 |
- Paal K, Ito M, and Withers SG. (2004). Paenibacillus sp. TS12 glucosylceramidase: kinetic studies of a novel sub-family of family 3 glycosidases and identification of the catalytic residues. Biochem J. 2004;378(Pt 1):141-9. DOI:10.1042/BJ20031028 |
- Bacik JP, Whitworth GE, Stubbs KA, Vocadlo DJ, and Mark BL. (2012). Active site plasticity within the glycoside hydrolase NagZ underlies a dynamic mechanism of substrate distortion. Chem Biol. 2012;19(11):1471-82. DOI:10.1016/j.chembiol.2012.09.016 |
- Vocadlo DJ, Mayer C, He S, and Withers SG. (2000). Mechanism of action and identification of Asp242 as the catalytic nucleophile of Vibrio furnisii N-acetyl-beta-D-glucosaminidase using 2-acetamido-2-deoxy-5-fluoro-alpha-L-idopyranosyl fluoride. Biochemistry. 2000;39(1):117-26. DOI:10.1021/bi991958d |
- Vocadlo DJ and Withers SG. (2005). Detailed comparative analysis of the catalytic mechanisms of beta-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry. 2005;44(38):12809-18. DOI:10.1021/bi051121k |
- Stubbs KA, Balcewich M, Mark BL, and Vocadlo DJ. (2007). Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated beta-lactam resistance. J Biol Chem. 2007;282(29):21382-91. DOI:10.1074/jbc.M700084200 |
- Bohlin C, Praestgaard E, Baumann MJ, Borch K, Praestgaard J, Monrad RN, and Westh P. (2013). A comparative study of hydrolysis and transglycosylation activities of fungal β-glucosidases. Appl Microbiol Biotechnol. 2013;97(1):159-69. DOI:10.1007/s00253-012-3875-9 |
- Litzinger S, Fischer S, Polzer P, Diederichs K, Welte W, and Mayer C. (2010). Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism. J Biol Chem. 2010;285(46):35675-84. DOI:10.1074/jbc.M110.131037 |
- Henrissat B (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280 ( Pt 2)(Pt 2):309-16. DOI:10.1042/bj2800309 |
- Hrmova M, Varghese JN, De Gori R, Smith BJ, Driguez H, and Fincher GB. (2001). Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase. Structure. 2001;9(11):1005-16. DOI:10.1016/s0969-2126(01)00673-6 |
- Hrmova M, De Gori R, Smith BJ, Fairweather JK, Driguez H, Varghese JN, and Fincher GB. (2002). Structural basis for broad substrate specificity in higher plant beta-D-glucan glucohydrolases. Plant Cell. 2002;14(5):1033-52. DOI:10.1105/tpc.010442 |
- Nakatani Y, Cutfield SM, Cowieson NP, and Cutfield JF. (2012). Structure and activity of exo-1,3/1,4-β-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain. FEBS J. 2012;279(3):464-78. DOI:10.1111/j.1742-4658.2011.08439.x |
- Yoshida E, Hidaka M, Fushinobu S, Koyanagi T, Minami H, Tamaki H, Kitaoka M, Katayama T, and Kumagai H. (2010). Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 β-glucosidase from Kluyveromyces marxianus. Biochem J. 2010;431(1):39-49. DOI:10.1042/BJ20100351 |
- Li H, Zhao G, Miyake H, Umekawa H, Kimura T, Ohmiya K, and Sakka K. (2006). Identification of a catalytic residue of Clostridium paraputrificum N-acetyl-beta-D-glucosaminidase Nag3A by site-directed mutagenesis. Biosci Biotechnol Biochem. 2006;70(5):1127-33. DOI:10.1271/bbb.70.1127 |
- Zmudka MW, Thoden JB, and Holden HM. (2013). The structure of DesR from Streptomyces venezuelae, a β-glucosidase involved in macrolide activation. Protein Sci. 2013;22(7):883-92. DOI:10.1002/pro.2204 |
- Balcewich MD, Stubbs KA, He Y, James TW, Davies GJ, Vocadlo DJ, and Mark BL. (2009). Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ. Protein Sci. 2009;18(7):1541-51. DOI:10.1002/pro.137 |
- Yamaguchi T, Blázquez B, Hesek D, Lee M, Llarrull LI, Boggess B, Oliver AG, Fisher JF, and Mobashery S. (2012). Inhibitors for Bacterial Cell-Wall Recycling. ACS Med Chem Lett. 2012;3(3):238-242. DOI:10.1021/ml2002746 |
- Vötsch W and Templin MF. (2000). Characterization of a beta -N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and beta -lactamase induction. J Biol Chem. 2000;275(50):39032-8. DOI:10.1074/jbc.M004797200 |
- Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, and Mark BL. (2009). Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(6):2274-82. DOI:10.1128/AAC.01617-08 |
- Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson KS, and Vorgias CE. (1996). Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol. 1996;3(7):638-48. DOI:10.1038/nsb0796-638 |
- Mark BL, Vocadlo DJ, Knapp S, Triggs-Raine BL, Withers SG, and James MN. (2001). Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J Biol Chem. 2001;276(13):10330-7. DOI:10.1074/jbc.M011067200 |
- Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, and Davies GJ. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol. 2006;13(4):365-71. DOI:10.1038/nsmb1079 |