CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 3"
| Line 18: | Line 18: | ||
CBM3 is a Gram-positive bacterial family of protein modules that comprise around 150 amino acids. The family is divided into four subgroups, CBM3a-d. The major ligand recognised by CBM3as and CBM3bs is crystalline cellulose with an affinity (K<sub>D</sub>) of 0.4 µM determined by depletion isotherms <cite>Morag1995</cite>. Isothermal titration calorimetry showed that binding to crystalline cellulose was entropically driven consistent with apolar interactions resulting in the release of caged water molecules from a ligand with a restricted conformation <cite>Hernandez-Gomez2015</cite>. CBM3s that bind to crystalline cellulose also interact with chitin and xyloglucan with an affinity ~500 lower than for crystalline cellulose. | CBM3 is a Gram-positive bacterial family of protein modules that comprise around 150 amino acids. The family is divided into four subgroups, CBM3a-d. The major ligand recognised by CBM3as and CBM3bs is crystalline cellulose with an affinity (K<sub>D</sub>) of 0.4 µM determined by depletion isotherms <cite>Morag1995</cite>. Isothermal titration calorimetry showed that binding to crystalline cellulose was entropically driven consistent with apolar interactions resulting in the release of caged water molecules from a ligand with a restricted conformation <cite>Hernandez-Gomez2015</cite>. CBM3s that bind to crystalline cellulose also interact with chitin and xyloglucan with an affinity ~500 lower than for crystalline cellulose. | ||
| − | Interaction with soluble xyloglucans by CBM3s was enthalpically driven with changes in entropy having a negative impact on affinity <cite>Hernandez-Gomez2015</cite>. The site of binding of a CBM3 from the ''Clostridium thermocellulum'' scaffoldin (CipA) to crystalline cellulose was determined by transmission electron microscopy with detection of the protein by immuno-gold labelling. The data showed that the CBM3 bound to the 110 face of Valonia cellulose <cite>Lehtio2003</cite>. The binding profile and site of cellulose recognition show that CBM3s are type A modules. The three CBM3s from anti-sigma sensors displayed different specificities; Cthe_0059 CBM3b bound to a range of plant cell wall polysaccharides (PCWPs), Cthe_0404 CBM3b interacted weakly to xyloglucan but not to any other PCWP, and Cthe_0267 CBM3 bound primarily to crystalline and amorphous cellulose <cite>Nataf2010,Yaniv2014</cite>. | + | Interaction with soluble xyloglucans by CBM3s was enthalpically driven with changes in entropy having a negative impact on affinity <cite>Hernandez-Gomez2015</cite>. The site of binding of a CBM3 from the ''Clostridium thermocellulum'' scaffoldin (CipA) to crystalline cellulose was determined by transmission electron microscopy with detection of the protein by immuno-gold labelling. The data showed that the CBM3 bound to the 110 face of Valonia cellulose <cite>Lehtio2003</cite>. The binding profile and site of cellulose recognition show that CBM3s are [[Carbohydrate-binding_modules#Types|type A]] modules. The three CBM3s from anti-sigma sensors displayed different specificities; Cthe_0059 CBM3b bound to a range of plant cell wall polysaccharides (PCWPs), Cthe_0404 CBM3b interacted weakly to xyloglucan but not to any other PCWP, and Cthe_0267 CBM3 bound primarily to crystalline and amorphous cellulose <cite>Nataf2010,Yaniv2014</cite>. |
== Structural Features == | == Structural Features == | ||
Revision as of 08:00, 6 March 2018
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Harry Gilbert^^^ and ^^^Ed Bayer^^^
- Responsible Curator: ^^^Ed Bayer^^^
| CAZy DB link | |
| https://www.cazy.org/CBM3.html |
Ligand specificities
CBM3 is a Gram-positive bacterial family of protein modules that comprise around 150 amino acids. The family is divided into four subgroups, CBM3a-d. The major ligand recognised by CBM3as and CBM3bs is crystalline cellulose with an affinity (KD) of 0.4 µM determined by depletion isotherms [1]. Isothermal titration calorimetry showed that binding to crystalline cellulose was entropically driven consistent with apolar interactions resulting in the release of caged water molecules from a ligand with a restricted conformation [2]. CBM3s that bind to crystalline cellulose also interact with chitin and xyloglucan with an affinity ~500 lower than for crystalline cellulose.
Interaction with soluble xyloglucans by CBM3s was enthalpically driven with changes in entropy having a negative impact on affinity [2]. The site of binding of a CBM3 from the Clostridium thermocellulum scaffoldin (CipA) to crystalline cellulose was determined by transmission electron microscopy with detection of the protein by immuno-gold labelling. The data showed that the CBM3 bound to the 110 face of Valonia cellulose [3]. The binding profile and site of cellulose recognition show that CBM3s are type A modules. The three CBM3s from anti-sigma sensors displayed different specificities; Cthe_0059 CBM3b bound to a range of plant cell wall polysaccharides (PCWPs), Cthe_0404 CBM3b interacted weakly to xyloglucan but not to any other PCWP, and Cthe_0267 CBM3 bound primarily to crystalline and amorphous cellulose [4, 5].
Structural Features
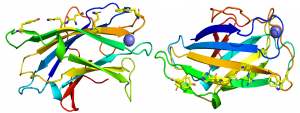
The crystal structure of CBM3 from the C. thermocellum scaffoldin CipA revealed a classical β-jelly-roll fold consisting of nine β-strands in two antiparallel β-sheets comprising four (1, 2, 7, 4; β-sheet 1) and five (9, 8, 3, 6, 5; β-sheet 2) β strands, respectively [6] (PDB ID 1NBC). β-sheet 1 forms a flat surface that contains a linear array of five residues that presents a planar hydrophobic surface comprising a His, Trp, Tyr and an Arg-Asp ion pair (Figure 1). The residues in the planar strip were predicted to make apolar interactions with glucose molecules n, n+1, n+3 and n+5, consistent with mutagenesis data showing that each of the five amino acids played an important role in binding cellulose [7]. Structures of CBMbs from other cellulosome-producing species followed that reinforced the original structural findings [8, 9, 10].
In other CBM3 modules that bind to cellulose, such as in the anti-σ-cell surface sensor RsgI1 (Cthe_0059), the His and ion pair are replaced by a Tyr and Phe, thus the hydrophobic planar binding site comprises four aromatic amino acids [5]. In a second cellulose-binding CBM3 located in an Rsgl sensor (Rsgl2, Cthe_0267), the aromatic planar strip is truncated, but lies planar with a hydrophobic protruding loop that is predicted to contribute to the cellulose binding site of the protein, similar to a group d CBM3 present in a GH48 exo-cellulase [11]. In addition to the hydrophobic strips it has also been proposed that highly conserved polar residues may be able to make productive hydrogen bonds with two additional cellulose chains in the microfibril [5, 6].
In contrast to the planar face presented by β-sheet 1, β-sheet 2 displays a concave surface or shallow groove that contains highly conserved aromatic residues [6], suggesting that these hydrophobic amino acids are functionally significant. It has been proposed that the shallow cleft is involved in binding Pro-Thr linker segments and thus may contribute to structural organization of these multimodular proteins [12].
Functionalities
CBM3s are derived from the scaffoldins [13] (non-catalytic proteins that that play an integral role in the assembly of multienzyme plant cell wall degrading complexes termed cellulosomes (see [14] for review), sensor proteins that detect cellulose [4] and a range of cellulases (e.g. [15, 16, 17]). In general CBM3s are separated from the other modules in these proteins by Pro-Thr-rich linker sequences. In some instances, however, group c CBM3 members (CBM3cs) are integral components of the substrate binding cleft of GH9 cellulases (e.g. [16, 17, 18, 19]) In these enzymes the CBM3c modules, as discrete entities, do not bind to cellulose (reflecting the lack of conserved ligand binding residues) but play a pivotal role in the capacity of the cellulases to attack crystalline forms of the polysaccharide [17]. It was proposed that the replacement of aromatic residues with conserved polar amino acids, altered the function of CBM3cs from an anchoring role. In the model proposed the polar residues in CBM3s replace the inter-chain hydrogen bonds within crystalline cellulose. The resultant disruption of the crystalline polysaccharide releases the cellulose chain on the centre of the CBM3c, which could then be fed into the active site cleft of the catalytic domain. Several studies have shown that CBM3 modules have enhanced the activity of cellulases [20] and a range of other plant cell wall degrading enzymes [21, 22]. These modules have also been used to probe the structure of plant cell walls [23, 24].
Family Firsts
- First Identified
- The first CBM3 to be identified (CipA-CBM3) was from the C. thermocellum scaffoldin CipA [13].
- First Structural Characterization
- The first crystal structure of a CBM3, indeed of any CBM, is CipA-CBM3 [6]
References
Error fetching PMID 8918451:
Error fetching PMID 11092922:
Error fetching PMID 21636890:
Error fetching PMID 22232162:
Error fetching PMID 24531486:
Error fetching PMID 21803997:
Error fetching PMID 20373916:
Error fetching PMID 20937888:
Error fetching PMID 24357319:
Error fetching PMID 12511483:
Error fetching PMID 9334746:
Error fetching PMID 22608730:
Error fetching PMID 16445761:
Error fetching PMID 19302786:
Error fetching PMID 12397074:
Error fetching PMID 24297170:
Error fetching PMID 17068959:
- Morag E, Lapidot A, Govorko D, Lamed R, Wilchek M, Bayer EA, and Shoham Y. (1995). Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl Environ Microbiol. 1995;61(5):1980-6. DOI:10.1128/aem.61.5.1980-1986.1995 |
- Hernandez-Gomez MC, Rydahl MG, Rogowski A, Morland C, Cartmell A, Crouch L, Labourel A, Fontes CM, Willats WG, Gilbert HJ, and Knox JP. (2015). Recognition of xyloglucan by the crystalline cellulose-binding site of a family 3a carbohydrate-binding module. FEBS Lett. 2015;589(18):2297-303. DOI:10.1016/j.febslet.2015.07.009 |
- Error fetching PMID 12522267:
- Error fetching PMID 20937888:
- Error fetching PMID 24531486:
- Error fetching PMID 8918451:
-
Benhar, I., Tamarkin, A., Marash, L., Berdichevsky, Y., Yaron, S., Shoham, Y., Lamed, R., and Bayer, E. A. (2001) Phage display of cellulose binding domains for biotechnological application. In Glycosyl Hydrolases for Biomass Conversion (Himmel, M. E., Baker, J. O., and Saddler, J. N., Eds.), pp 168-189, American Chemical Society, Washington, DC. DOI:10.1021/bk-2001-0769.ch010.
- Error fetching PMID 11092922:
- Error fetching PMID 21636890:
- Error fetching PMID 22232162:
- Error fetching PMID 21803997:
- Error fetching PMID 22608730:
- Poole DM, Morag E, Lamed R, Bayer EA, Hazlewood GP, and Gilbert HJ. (1992). Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol Lett. 1992;78(2-3):181-6. DOI:10.1016/0378-1097(92)90022-g |
- Error fetching PMID 20373916:
- Error fetching PMID 24357319:
- Error fetching PMID 12511483:
- Error fetching PMID 9334746:
- Error fetching PMID 16445761:
- Error fetching PMID 19302786:
- Error fetching PMID 12397074:
- Error fetching PMID 24297170:
- Hervé C, Rogowski A, Blake AW, Marcus SE, Gilbert HJ, and Knox JP. (2010). Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A. 2010;107(34):15293-8. DOI:10.1073/pnas.1005732107 |
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, and Knox JP. (2006). Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem. 2006;281(39):29321-9. DOI:10.1074/jbc.M605903200 |
- Error fetching PMID 17068959: