CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 101"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) |
||
| Line 32: | Line 32: | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | |||
Using the enzyme from ''Streptococcus pneumoniae'' the [[catalytic nucleophile]] was determined as residue D764. Willis ''et al.'' in preparation <cite>5</cite> | Using the enzyme from ''Streptococcus pneumoniae'' the [[catalytic nucleophile]] was determined as residue D764. Willis ''et al.'' in preparation <cite>5</cite> | ||
The [[general acid/base]] residue in SpGH101 was determined to be E796. | The [[general acid/base]] residue in SpGH101 was determined to be E796. | ||
| − | |||
== Three-dimensional structures == | == Three-dimensional structures == | ||
Revision as of 01:21, 28 September 2009
| Glycoside Hydrolase Family GH101 | |
| Clan | GH-x |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/fam/GH101.html | |
Substrate specificities
Glycoside hydrolase family 101 currently contains proteins from bacterial species, most of which are commensal human bacteria, though some may also be human pathogens. The substrates are glycoproteins which contain the disaccharide Gal-beta-1,3-GalNAc-alpha-R also know as the Core 1 O-linked glycans on proteins. This glycosylation is a feature of mucin proteins. This enzyme activity was first observed in Clostridium perfringens [1] and then in Streptococcus pnuemoniae [2]. At present the known enzymes will not digest longer oligosaccharides, and variable amounts of activity have been seen on Core 2 and 3 type linkages [3].
Kinetics and Mechanism
A retaining mechanism was determined by 1H-NMR with the BlGH101 enzyme [4]. A detailed kinetic study has been performed with the SpGH101 by Willis and co-workers and the catalytic residues have been determined [5].
Catalytic Residues
Using the enzyme from Streptococcus pneumoniae the catalytic nucleophile was determined as residue D764. Willis et al. in preparation [5] The general acid/base residue in SpGH101 was determined to be E796.
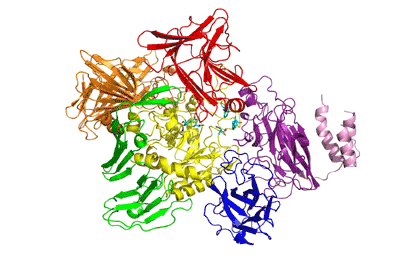
Three-dimensional structures
The recently solved 3D structure of SpGH101 reveals that this protein surprisingly shares some structural features with GH13 alpha-amylases, as well as containing potential carbohydrate binding modules. In the analysis of the 3D structure of SpGH101 the catalytic domain is a highly distorted (beta/alpha)8 barrel. This barrel is flanked by a domain of all beta-sheet structure which is very analogous to the arrangement seen in GH13 alpha-amylase [6]. The BlGH101 enzyme was found to have an almost identical fold, although this structure was solved at higher resolution (2A) than that of the SpGH101 (2.9A) [7]
Family Firsts
- First sterochemistry determination
- This was determined with the BlGH101 enzyme using the 1H-NMR technique with the synthetic substrate Gal-beta-1,3-GalNAc-p-nitrophenol [4].
- First catalytic nucleophile identification
This was proposed based on the structure of the SpGH101 and BlGH101 structures, and then experimentally shown in SpGH101 by Willis and co-workers [5].
- First general acid/base residue identification
experimentally shown in SpGH101 by Willis and co-workers [5]
- First 3-D structure
Determined for SpGH101 by Caines and co-workers [6]
References
- Huang CC and Aminoff D. (1972). Enzymes that destroy blood group specificity. V. The oligosaccharase of Clostridium perfringens. J Biol Chem. 1972;247(21):6737-42. | Google Books | Open Library
- Bhavanandan VP, Umemoto J, and Davidson EA. (1976). Characterization of an endo-alpha-N-acetyl galactosaminidase from Diplococcus pneumoniae. Biochem Biophys Res Commun. 1976;70(3):738-45. DOI:10.1016/0006-291x(76)90654-9 |
- Koutsioulis D, Landry D, and Guthrie EP. (2008). Novel endo-alpha-N-acetylgalactosaminidases with broader substrate specificity. Glycobiology. 2008;18(10):799-805. DOI:10.1093/glycob/cwn069 |
- Fujita K, Oura F, Nagamine N, Katayama T, Hiratake J, Sakata K, Kumagai H, and Yamamoto K. (2005). Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem. 2005;280(45):37415-22. DOI:10.1074/jbc.M506874200 |
-
in preparation
- Caines ME, Zhu H, Vuckovic M, Willis LM, Withers SG, Wakarchuk WW, and Strynadka NC. (2008). The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design. J Biol Chem. 2008;283(46):31279-83. DOI:10.1074/jbc.C800150200 |
- Suzuki R, Katayama T, Kitaoka M, Kumagai H, Wakagi T, Shoun H, Ashida H, Yamamoto K, and Fushinobu S. (2009). Crystallographic and mutational analyses of substrate recognition of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J Biochem. 2009;146(3):389-98. DOI:10.1093/jb/mvp086 |