CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 48"
| Line 51: | Line 51: | ||
Three-dimensional structures are available for two family 48 enzymes: Cel48F (from Clostridium cellulolyticum) and Cel48A (from Clostridium thermocellum). Both enzymes have an (α/α)6 barrel topology. | Three-dimensional structures are available for two family 48 enzymes: Cel48F (from Clostridium cellulolyticum) and Cel48A (from Clostridium thermocellum). Both enzymes have an (α/α)6 barrel topology. | ||
[[Image:1FAE.jpg|thumb| left| ]] | [[Image:1FAE.jpg|thumb| left| ]] | ||
| − | + | [[Image:1L1Y.jpg|thumb| right| PDB:1L1Y ]] | |
[http://www.rcsb.org/pdb/explore/explore.do?structureId=1FAE Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose] | [http://www.rcsb.org/pdb/explore/explore.do?structureId=1FAE Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose] | ||
Revision as of 12:05, 14 February 2010
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Bareket Dassa^^^
- Responsible Curator: ^^^Ed Bayer^^^
| Glycoside Hydrolase Family GH48 | |
| Clan | GH-M |
| Mechanism | inverting |
| Active site residues | not known |
| CAZy DB link | |
| http://www.cazy.org/fam/GH48.html | |
Family 48 glycoside hydrolases are major and key components of some cellulase systems, occurring in free enzyme systems (e.g., in Thermobifida fusca), multi-functional enzymes (e.g, in Caldocellulosiruptor saccharolyticus), anaerobic fungi (e.g., Piromyces equi) and every cellulosome system thus far described. The GH48 cellulase is commonly the most abundant enzyme subunit in cellulosome-producing bacteria. Each bacterium usually contains a single gene that codes for a GH48 enzyme, although a few bacteria (e.g., and Anaerocellum thermophilum) contain two or more GH48 genes. Of the two C. thermocellum GH48 enzymes, one (Cel48S) is a dockerin-containing cellulosomal enzyme, and the other (Cel48Y) is a free, non-cellulosomal enzyme that contains a cellulose-binding CBM3.
Substrate specificities
The following activities have been reported: endo-b-1,4-glucanase, chitinase, endo-processive cellulase and cellobiohydrolase. Its preferred substrate is amorphous or crystalline cellulose over carboxymethylcellulose (CMC), and its activity is strongly inhibited by the presence of cellobiose. Although its activity on various substrates is characteristically very low, it is thought to be a critically important enzyme which imparts a major component of synergy to its cellulase system.
Kinetics and Mechanism
The glycoside hydrolases of this family includes inverting glycosidases, which preferentially attack the reducing end of the substrate [1].
The native and recombinant Cel48S from C. thermocellum displays typical characteristics of a processive exoglucanase [2], and its activity on amorphous cellulose is optimal at 70 °C and at pH 5–6.
Family 48 cellulases (i.e., CelS/S8 from C. thermocellum, Avicelase II of C. stercorarium) are stabilized at high temperatures by Ca2+ or other bivalent ions [3, 4, 5].
The Cel48F protein from Clostridium cellulolyticum has been reported [6] to be a processive endo-glucanase, which performs a processive degradation of the cellulose chain after an initial endo-attack. A two-step mechanism was proposed [7], in which processive action and chain disruption occupy different subsites.
Catalytic Residues
The crystal structure of Cel48F, a cellulosome component of C. cellulolyticum, revealed the active center at the junction of the cleft and tunnel regions, where Glu55 is the proposed proton donor in the cleavage reaction, and the corresponding base was initially proposed to be either Glu44 or Asp230 [8].
The structure of the catalytic module of Cel48S of C. thermocellum showed a similar tunnel-shaped substrate-binding region formed by the alpha helices in the protein. The hydrolysis of the cellulose chain in Cel48S appeared to involve Glu87 (the equivalent of Glu55 in C. cellulolyticum Cel48F) as an acid to protonate the glycosidic oxygen atom and Tyr351 as a base to extract a proton from the nucleophilic water molecule that attacks the anomeric carbon atom.
More recent studies of Cel48F failed to unambiguously identity the catalytic base in the cleavage reaction [7].
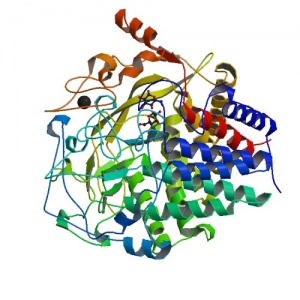
Three-dimensional structures
Three-dimensional structures are available for two family 48 enzymes: Cel48F (from Clostridium cellulolyticum) and Cel48A (from Clostridium thermocellum). Both enzymes have an (α/α)6 barrel topology.
Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose
The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome [9]
Family Firsts
- First sterochemistry determination
- Cellulomonas fimi CenE, described as an endo-β-1,4-glucanase, catalyzes the hydrolysis of cellohexaose with inversion of anomeric carbon configuration, characteristic of a single displacement reaction [10].
- First catalytic nucleophile identification
- …“Waiting patiently”… (see [7]).
- First general acid/base residue identification
- First 3-D structure
- The crystal structure of catalytic module of C. cellulolyticum Cel48F in complex with oligosaccharides [7].
- First cloning and sequencing
- The cel48S gene from C. thermocellum [11].
References
- Katosova LK (1977). [Cytochemical characteristics of blood leukocytes during an experimental purulent-inflammatory process in rabbits]. Zh Mikrobiol Epidemiol Immunobiol. 1977(3):102-7. | Google Books | Open Library
- Guimarães BG, Souchon H, Lytle BL, David Wu JH, and Alzari PM. (2002). The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome. J Mol Biol. 2002;320(3):587-96. DOI:10.1016/s0022-2836(02)00497-7 |
- Bronnenmeier K, Rücknagel KP, and Staudenbauer WL. (1991). Purification and properties of a novel type of exo-1,4-beta-glucanase (avicelase II) from the cellulolytic thermophile Clostridium stercorarium. Eur J Biochem. 1991;200(2):379-85. DOI:10.1111/j.1432-1033.1991.tb16195.x |
- Morag E, Halevy I, Bayer EA, and Lamed R. (1991). Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J Bacteriol. 1991;173(13):4155-62. DOI:10.1128/jb.173.13.4155-4162.1991 |
- Kruus K, Wang WK, Ching J, and Wu JH. (1995). Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J Bacteriol. 1995;177(6):1641-4. DOI:10.1128/jb.177.6.1641-1644.1995 |
- Reverbel-Leroy C, Pages S, Belaich A, Belaich JP, and Tardif C. (1997). The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J Bacteriol. 1997;179(1):46-52. DOI:10.1128/jb.179.1.46-52.1997 |
- Parsiegla G, Reverbel C, Tardif C, Driguez H, and Haser R. (2008). Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action. J Mol Biol. 2008;375(2):499-510. DOI:10.1016/j.jmb.2007.10.039 |
- Parsiegla G, Juy M, Reverbel-Leroy C, Tardif C, Belaïch JP, Driguez H, and Haser R. (1998). The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 A resolution. EMBO J. 1998;17(19):5551-62. DOI:10.1093/emboj/17.19.5551 |
- Guimarães BG, Souchon H, Lytle BL, David Wu JH, and Alzari PM. (2002). The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome. J Mol Biol. 2002;320(3):587-96. DOI:10.1016/s0022-2836(02)00497-7 |
- Shen H, Tomme P, Meinke A, Gilkes NR, Kilburn DG, Warren RA, and Miller RC Jr. (1994). Stereochemical course of hydrolysis catalysed by Cellulomonas fimi CenE, a member of a new family of beta-1,4-glucanases. Biochem Biophys Res Commun. 1994;199(3):1223-8. DOI:10.1006/bbrc.1994.1361 |
- Wang WK, Kruus K, and Wu JH. (1993). Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase Ss (CelS), a major cellulosome component. J Bacteriol. 1993;175(5):1293-302. DOI:10.1128/jb.175.5.1293-1302.1993 |