CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Cellulosome"
| Line 32: | Line 32: | ||
* ''Clostridium thermocellum'' <cite>species9</cite> | * ''Clostridium thermocellum'' <cite>species9</cite> | ||
* ''Ruminococcus albus'' (dockerins identified, cohesins as yet undetected) <cite>species10</cite> | * ''Ruminococcus albus'' (dockerins identified, cohesins as yet undetected) <cite>species10</cite> | ||
| − | * ''Ruminococcus | + | * ''Ruminococcus flavefaciens'' <cite>species11 species12</cite> |
Cellulosomes exist as extracellular complexes that are either attached to the cell wall of bacteria or free in solution, where the insoluble substrate can be broken down into soluble products and taken up by the cell. The large size and heterogeneity of cellulosomes from the best-characterized organisms (i.e., ''C. thermocellum'', ''C. cellulolyticum'', and ''C. cellulovorans'') have greatly complicated efforts to probe cellulosome structure and function. Other cellulosome systems (such as those from ''Acetivibrio cellulolyticus'' and ''Ruminococcus flavefaciens'') appear to be even more intricate. | Cellulosomes exist as extracellular complexes that are either attached to the cell wall of bacteria or free in solution, where the insoluble substrate can be broken down into soluble products and taken up by the cell. The large size and heterogeneity of cellulosomes from the best-characterized organisms (i.e., ''C. thermocellum'', ''C. cellulolyticum'', and ''C. cellulovorans'') have greatly complicated efforts to probe cellulosome structure and function. Other cellulosome systems (such as those from ''Acetivibrio cellulolyticus'' and ''Ruminococcus flavefaciens'') appear to be even more intricate. | ||
Revision as of 03:10, 16 June 2010
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Authors: ^^^Orly Alber^^^, ^^^Bareket Dassa^^^, and ^^^Ed Bayer^^^
- Responsible Curator: ^^^Ed Bayer^^^
Cellulosome complex
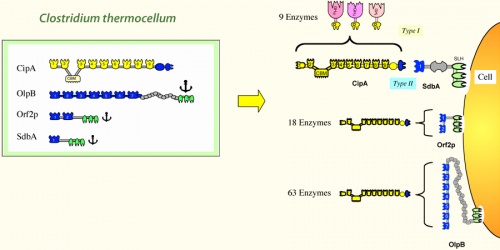
Cellulosome complexes are intricate multi-enzyme machines produced by many cellulolytic microorganisms. They are designed for efficient degradation of plant cell wall polysaccharides, notably cellulose — the most abundant organic polymer on Earth. The cellulosome consists of a multi-functional integrating subunit (called scaffoldin), responsible for organizing the various cellulolytic subunits (e.g., the enzymes) into the complex. Within a cellulosome, multiple endoglucanases, cellobiohydrolases, xylanases and other degradative enzymes work synergistically to attack heterogeneous, insoluble cellulose substrates. This is accomplished by the interaction of two complementary classes of module, located on the two separate types of interacting subunits, i.e., a cohesin module on the scaffoldin and a dockerin module on each enzymatic subunit. The high-affinity cohesin-dockerin interaction defines the cellulosome structure. Attachment of the cellulosome to its substrate is mediated by a scaffoldin-borne cellulose-binding module (CBM) that comprises part of the scaffoldin subunit. Much of our understanding of its catalytic components, architecture, and mechanisms of attachment to the bacterial cell and to cellulose, has been derived from the study of Clostridium thermocellum.
Cellulosome components:
- The scaffoldin subunit contains one or more cohesin modules connected to other types of functional modules. In a given scaffoldin, the latter types of modules may include a cellulose-specific carbohydrate-binding module (CBM), a dockerin, X modules of unknown function, an S-layer homology (SLH) module or a sortase anchoring motif.
- Cohesin modules are the major building blocks of scaffoldins, which are responsible for organizing the cellulolytic subunits into the multi-enzyme complex.
- Dockerin modules anchor the catalytic enzymes to the scaffoldin. The dockerin displays internal two-fold symmetry, consisting of a duplicated F-hand motif (a calcium-binding loop preceding an a helix). The dockerin can also be found in the C- terminus of scaffoldins.
- Catalytic subunits contain dockerin modules that serve to incorporate catalytic modules into the cellulosome complex. These catalytic modules include: glycoside hydrolases, polysaccharide lyases, and carboxyl esterases (See also [1, 2, 3]).
Cellulosome systems
Early on [4] it became clear that cellulosomes were not restricted to C. thermocellum, but were present in other cellulolytic bacteria. Bacterial cellulosomal systems can be categorized into two major types: simple cellulosome systems contain a single scaffoldin and complex cellulosome systems exhibit multiple types of interacting scaffoldins. The arrangement of the modules on the scaffoldin subunit and the specificity of the cohesin(s) and/or dockerin for their modular counterpart dictate the overall architecture of the cellulosome. Several different types of scaffoldins have been described: the primary scaffoldins incorporate the various dockerin-bearing subunits directly into the cellulosome complex, adaptor scaffoldins increase the repertoire or number of components into the complex, and the anchoring scaffoldins attach the complex to the bacterial cell surface.
Currently known cellulosome-producing anaerobic bacteria:
- Acetivibrio cellulolyticus [5]
- Bacteroides cellulosolvens [6]
- Clostridium acetobutylicum [7]
- Clostridium cellobioparum (suspected, not proven) [8]
- Clostridium cellulolyticum [9]
- Clostridium cellulovorans [10]
- Clostridium josui [11]
- Clostridium papyrosolvens [12]
- Clostridium thermocellum [13]
- Ruminococcus albus (dockerins identified, cohesins as yet undetected) [14]
- Ruminococcus flavefaciens [15, 16]
Cellulosomes exist as extracellular complexes that are either attached to the cell wall of bacteria or free in solution, where the insoluble substrate can be broken down into soluble products and taken up by the cell. The large size and heterogeneity of cellulosomes from the best-characterized organisms (i.e., C. thermocellum, C. cellulolyticum, and C. cellulovorans) have greatly complicated efforts to probe cellulosome structure and function. Other cellulosome systems (such as those from Acetivibrio cellulolyticus and Ruminococcus flavefaciens) appear to be even more intricate.
Simple Cellulosome Systems In the simple cellulosome systems, the scaffoldins contain a single CBM, one or more X2 modules and numerous (5 to 9) cohesins. These scaffoldins are primary scaffoldins, which incorporate the dockerin-bearing enzymes into the complex. In several cases, the simple cellulosomes have been shown to be associated with the cell surface, but the molecular mechanism responsible for this is still unclear. The X2 module may play a role in attachment to the cell wall. The genes encoding for many important cellulosome subunits are organized in “enzyme-linked gene clusters” on the chromosome.
Complex Cellulosome Systems To date, complex cellulosome systems have been described in different bacterial species (See also [2, 3, 17]). In these systems, more than one scaffoldin interlocks with each other in various ways to produce a complex cellulosome architecture. At least one type of scaffoldin serves as a primary scaffoldin that incorporates the enzymes directly into the cellulosome complex. In each species, another type of scaffoldin attaches the cellulosome complex to the cell surface via a specialized module or sequence, designed for this purpose. In the complex cellulosome systems, the scaffoldin genes are organized into “multiple scaffoldin gene clusters” on the chromosome.
Cohesin-dockerin interactions
Cohesin-dockerin interactions can be viewed as a kind of plug-and-socket mechanism in which the dockerin plugs into the cohesin socket1. In general, the interaction is inter-species and intra-species (type) specific, although some cross-reactivity has been found in a few cases. The cohesin-dockerin interaction is one of the most potent protein-protein interactions known in nature, in most cases approaching the strength of high-affinity antigen-antibody interactions (Ka ~ 1011 M-1).
So far, cohesins have been phylogenetically distributed into three groups according to sequence homology; the type-I cohesin, the type-II cohesin and the recently discovered type-III cohesin. The dockerins that interact with each cohesin type are, by definition, of the same type.
Structural characterization of cellulosome components
One of the greatest efforts in the cellulosome research field is to understand the structure-function relationship in cellulosome assembly. Thus far, crystallographic structures of only selected cohesins have been determined, all of which share the typical jelly-roll topology that forms a flattened 9-stranded beta-sandwich. In addition, crystal structures for type-I and type-II cohesin-dockerin complexes have been described. The structure of a multimodular complex from C. thermocellum was also solved, composed of the type-II cohesin module of the cell surface protein SdbA bound to a trimodular C-terminal fragment of the scaffoldin subunit CipA.

History of discovery
In the early 1980s, Raffi Lamed and Ed Bayer met at Tel Aviv University and commenced their work that led to the discovery of the cellulosome concept. At the time, they weren’t looking for enzymes or cellulosomes at all. They simply sought a ‘cellulose-binding factor’ or ‘CBF’ on the cell surface of the anaerobic thermophilic bacterium, C. thermocellum, which they inferred would account for the observation that the bacterium attaches strongly to the insoluble cellulose substrate prior to its degradation. They employed a then unconventional experimental approach, in which they isolated an adherence-defective mutant of the bacterium and prepared a specific polyclonal antibody for detection of the functional component. Surprisingly, they isolated a very large multi-subunit supramolecular complex, instead of a small protein. A combination of biochemical, biophysical, immunochemical and ultrastructural techniques, followed by molecular biological verification, led to the definition and proof of the cellulosome concept. The birth of the discrete, multi-enzyme cellulosome complex was thus documented.
Cellulosome Firsts
- Discovery of the cellulosome in Clostridium thermocellum [19, 20]
- Demonstration of true cellulase activity by the cellulosome (Lamed et al., Enzyme Microb Technol 1985;7:37-41, 1985)
- Detailed ultrastructural characterization of the cellulosome [21, 22]
- Demonstration of the cellulosome-like entities in other cellulose-degrading strains [23]
- Crystal structure of cellulosomal enzyme (Juy et al.,Nature 1992;357:39-41, 1992)
- Functional role of dockerin module (duplicated domain) [24]
- Sequencing and characterization of primary scaffoldin genes [25, 26]
- Sequencing of cell-surface anchoring scaffoldins [27]
- Definition of “cohesin”, “dockerin” and “scaffoldin” [28]
- Functional role of cohesins [29]
- Establishment of designer cellulosome concept [28, 29, 30]
- Crystal structures of cohesins and cohesin-dockerin complexes [30, 31, 32, 33]
Additional information is available on the Bayer lab website.
References
- Gilbert HJ (2007). Cellulosomes: microbial nanomachines that display plasticity in quaternary structure. Mol Microbiol. 2007;63(6):1568-76. DOI:10.1111/j.1365-2958.2007.05640.x |
- Demain AL, Newcomb M, and Wu JH. (2005). Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev. 2005;69(1):124-54. DOI:10.1128/MMBR.69.1.124-154.2005 |
- Lamed R, Naimark J, Morgenstern E, and Bayer EA. (1987). Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987;169(8):3792-800. DOI:10.1128/jb.169.8.3792-3800.1987 |
- Ding SY, Bayer EA, Steiner D, Shoham Y, and Lamed R. (1999). A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J Bacteriol. 1999;181(21):6720-9. DOI:10.1128/JB.181.21.6720-6729.1999 |
- Ding SY, Bayer EA, Steiner D, Shoham Y, and Lamed R. (2000). A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J Bacteriol. 2000;182(17):4915-25. DOI:10.1128/JB.182.17.4915-4925.2000 |
- Nölling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J, Wolf YI, Tatusov RL, Sabathe F, Doucette-Stamm L, Soucaille P, Daly MJ, Bennett GN, Koonin EV, and Smith DR. (2001). Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183(16):4823-38. DOI:10.1128/JB.183.16.4823-4838.2001 |
- Lamed R, Naimark J, Morgenstern E, and Bayer EA. (1987). Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987;169(8):3792-800. DOI:10.1128/jb.169.8.3792-3800.1987 |
- Pagès S, Bélaïch A, Fierobe HP, Tardif C, Gaudin C, and Bélaïch JP. (1999). Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol. 1999;181(6):1801-10. DOI:10.1128/JB.181.6.1801-1810.1999 |
- Shoseyov O, Takagi M, Goldstein MA, and Doi RH. (1992). Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc Natl Acad Sci U S A. 1992;89(8):3483-7. DOI:10.1073/pnas.89.8.3483 |
- Kakiuchi M, Isui A, Suzuki K, Fujino T, Fujino E, Kimura T, Karita S, Sakka K, and Ohmiya K. (1998). Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J Bacteriol. 1998;180(16):4303-8. DOI:10.1128/JB.180.16.4303-4308.1998 |
- Pohlschröder M, Canale-Parola E, and Leschine SB. (1995). Ultrastructural diversity of the cellulase complexes of Clostridium papyrosolvens C7. J Bacteriol. 1995;177(22):6625-9. DOI:10.1128/jb.177.22.6625-6629.1995 |
- Lamed R, Setter E, and Bayer EA. (1983). Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156(2):828-36. DOI:10.1128/jb.156.2.828-836.1983 |
- Lamed R, Naimark J, Morgenstern E, and Bayer EA. (1987). Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987;169(8):3792-800. DOI:10.1128/jb.169.8.3792-3800.1987 |
- Kirby J, Martin JC, Daniel AS, and Flint HJ. (1997). Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol Lett. 1997;149(2):213-9. DOI:10.1111/j.1574-6968.1997.tb10331.x |
- Ding SY, Rincon MT, Lamed R, Martin JC, McCrae SI, Aurilia V, Shoham Y, Bayer EA, and Flint HJ. (2001). Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J Bacteriol. 2001;183(6):1945-53. DOI:10.1128/JB.183.6.1945-1953.2001 |
- Adams JJ, Currie MA, Ali S, Bayer EA, Jia Z, and Smith SP. (2010). Insights into higher-order organization of the cellulosome revealed by a dissect-and-build approach: crystal structure of interacting Clostridium thermocellum multimodular components. J Mol Biol. 2010;396(4):833-9. DOI:10.1016/j.jmb.2010.01.015 |
- Bayer EA, Kenig R, and Lamed R. (1983). Adherence of Clostridium thermocellum to cellulose. J Bacteriol. 1983;156(2):818-27. DOI:10.1128/jb.156.2.818-827.1983 |
- Lamed R, Setter E, and Bayer EA. (1983). Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156(2):828-36. DOI:10.1128/jb.156.2.828-836.1983 |
- Bayer EA and Lamed R. (1986). Ultrastructure of the cell surface cellulosome of Clostridium thermocellum and its interaction with cellulose. J Bacteriol. 1986;167(3):828-36. DOI:10.1128/jb.167.3.828-836.1986 |
- Mayer F, Coughlan MP, Mori Y, and Ljungdahl LG. (1987). Macromolecular Organization of the Cellulolytic Enzyme Complex of Clostridium thermocellum as Revealed by Electron Microscopy. Appl Environ Microbiol. 1987;53(12):2785-92. DOI:10.1128/aem.53.12.2785-2792.1987 |
- Lamed R, Naimark J, Morgenstern E, and Bayer EA. (1987). Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987;169(8):3792-800. DOI:10.1128/jb.169.8.3792-3800.1987 |
- Tokatlidis K, Salamitou S, Béguin P, Dhurjati P, and Aubert JP. (1991). Interaction of the duplicated segment carried by Clostridium thermocellum cellulases with cellulosome components. FEBS Lett. 1991;291(2):185-8. DOI:10.1016/0014-5793(91)81279-h |
- Shoseyov O, Takagi M, Goldstein MA, and Doi RH. (1992). Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc Natl Acad Sci U S A. 1992;89(8):3483-7. DOI:10.1073/pnas.89.8.3483 |
- Gerngross UT, Romaniec MP, Kobayashi T, Huskisson NS, and Demain AL. (1993). Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol Microbiol. 1993;8(2):325-34. DOI:10.1111/j.1365-2958.1993.tb01576.x |
- Fujino T, Béguin P, and Aubert JP. (1993). Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J Bacteriol. 1993;175(7):1891-9. DOI:10.1128/jb.175.7.1891-1899.1993 |
- Bayer EA, Morag E, and Lamed R. (1994). The cellulosome--a treasure-trove for biotechnology. Trends Biotechnol. 1994;12(9):379-86. DOI:10.1016/0167-7799(94)90039-6 |
- Salamitou S, Raynaud O, Lemaire M, Coughlan M, Béguin P, and Aubert JP. (1994). Recognition specificity of the duplicated segments present in Clostridium thermocellum endoglucanase CelD and in the cellulosome-integrating protein CipA. J Bacteriol. 1994;176(10):2822-7. DOI:10.1128/jb.176.10.2822-2827.1994 |
- Fierobe HP, Mechaly A, Tardif C, Belaich A, Lamed R, Shoham Y, Belaich JP, and Bayer EA. (2001). Design and production of active cellulosome chimeras. Selective incorporation of dockerin-containing enzymes into defined functional complexes. J Biol Chem. 2001;276(24):21257-61. DOI:10.1074/jbc.M102082200 |
- Adams JJ, Pal G, Jia Z, and Smith SP. (2006). Mechanism of bacterial cell-surface attachment revealed by the structure of cellulosomal type II cohesin-dockerin complex. Proc Natl Acad Sci U S A. 2006;103(2):305-10. DOI:10.1073/pnas.0507109103 |
- Carvalho AL, Dias FM, Prates JA, Nagy T, Gilbert HJ, Davies GJ, Ferreira LM, Romão MJ, and Fontes CM. (2003). Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc Natl Acad Sci U S A. 2003;100(24):13809-14. DOI:10.1073/pnas.1936124100 |
- Tavares GA, Béguin P, and Alzari PM. (1997). The crystal structure of a type I cohesin domain at 1.7 A resolution. J Mol Biol. 1997;273(3):701-13. DOI:10.1006/jmbi.1997.1326 |
- Bayer EA, Belaich JP, Shoham Y, and Lamed R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521-54. DOI:10.1146/annurev.micro.57.030502.091022 |
- Fontes CM and Gilbert HJ. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem. 2010;79:655-81. DOI:10.1146/annurev-biochem-091208-085603 |
- Bayer EA, Lamed R, White BA, and Flint HJ. (2008). From cellulosomes to cellulosomics. Chem Rec. 2008;8(6):364-77. DOI:10.1002/tcr.20160 |
- Doi RH and Kosugi A. (2004). Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol. 2004;2(7):541-51. DOI:10.1038/nrmicro925 |
- Bayer EA, Morag E, and Lamed R. (1994). The cellulosome--a treasure-trove for biotechnology. Trends Biotechnol. 1994;12(9):379-86. DOI:10.1016/0167-7799(94)90039-6 |