CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Phosphorylases"
| Line 33: | Line 33: | ||
==Mechanism of glycoside hydrolase-like phosphorylases== | ==Mechanism of glycoside hydrolase-like phosphorylases== | ||
| − | Sequence and structural analysis of sugar phosphorylases reveal that | + | Sequence and structural analysis of sugar phosphorylases reveal that some have sequences and structures (and likely mechanisms) similar to [[glycosyltransferases]], whereas others have sequences and structures that more closely resemble [[glycoside hydrolase]]s. |
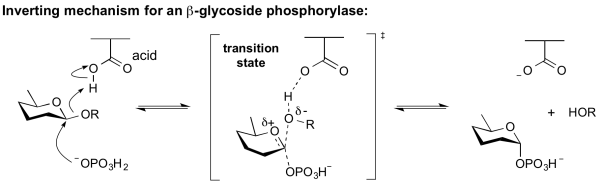
===Inverting phosphorylase mechanism=== | ===Inverting phosphorylase mechanism=== | ||
| − | Phosphorolysis of a glycoside by a GH-like phosphorylase with net inversion of anomeric configuration is achieved via a one step, single-displacement mechanism involving [[oxocarbenium ion]]-like [[transition state]]s, as shown below. The reaction occurs with acid/base assistance from a suitably positioned carboxylate residue. This mechanism has clear parallels with the mechanism for [[inverting]] glycoside hydrolases. | + | All inverting phosphorylases are GH-like in their sequence classification. Phosphorolysis of a glycoside by a GH-like phosphorylase with net inversion of anomeric configuration is achieved via a one step, single-displacement mechanism involving [[oxocarbenium ion]]-like [[transition state]]s, as shown below. The reaction occurs with acid/base assistance from a suitably positioned carboxylate residue. This mechanism has clear parallels with the mechanism for [[inverting]] glycoside hydrolases. |
<br><br> | <br><br> | ||
[[Image:Inverting_b-glycoside_phosphorylase_mechanism.png|center|600px]] | [[Image:Inverting_b-glycoside_phosphorylase_mechanism.png|center|600px]] | ||
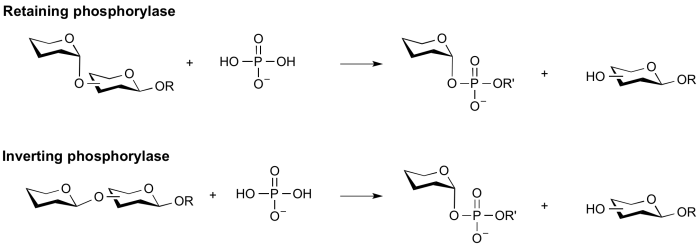
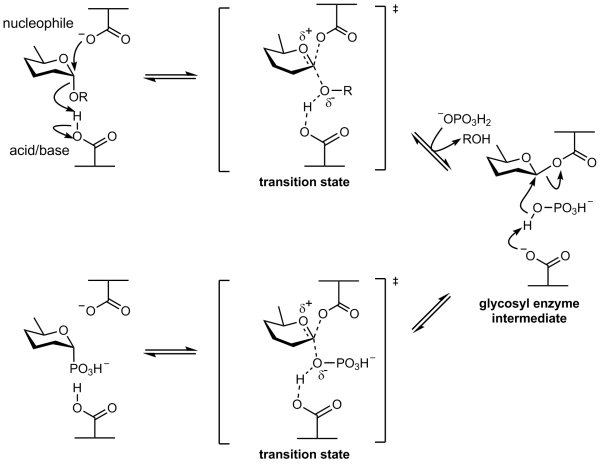
===Retaining phosphorylase mechanism=== | ===Retaining phosphorylase mechanism=== | ||
| − | Phosphorolysis of a glycoside with net retention of configuration is most probably achieved via a two step, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate, as shown in the figure below. Each step passes through an [[oxocarbenium ion]]-like [[transition state]]. Reaction occurs with acid/base and nucleophilic assistance provided by two amino acid side chains, typically glutamate or aspartate. In the first step (called the glycosylation step), one residue plays the role of a nucleophile, attacking the anomeric centre to displace the aglycon and form a glycosyl enzyme [[intermediate]]. At the same time the other residue functions as an acid catalyst and protonates the glycosidic oxygen as the bond cleaves. In the second step (known as the deglycosylation step), the glycosyl enzyme is hydrolyzed by phosphate, with the other residue now acting as a base catalyst deprotonating the phosphate as it attacks. An alternative mechanism for retention of anomeric configuration has been proposed involving direct front-side nucleophilic displacement (S<sub>N</sub>i). Both of these mechanisms bear considerable analogy to those proposed for retaining glycosyltransferases <cite>Lairsson2004 Lairsson2008 </cite>. | + | Phosphorolysis of a glycoside with net retention of configuration by the GH-like retaining phosphorylases is most probably achieved via a two step, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate, as shown in the figure below. Each step passes through an [[oxocarbenium ion]]-like [[transition state]]. Reaction occurs with acid/base and nucleophilic assistance provided by two amino acid side chains, typically glutamate or aspartate. In the first step (called the glycosylation step), one residue plays the role of a nucleophile, attacking the anomeric centre to displace the aglycon and form a glycosyl enzyme [[intermediate]]. At the same time the other residue functions as an acid catalyst and protonates the glycosidic oxygen as the bond cleaves. In the second step (known as the deglycosylation step), the glycosyl enzyme is hydrolyzed by phosphate, with the other residue now acting as a base catalyst deprotonating the phosphate as it attacks. An alternative mechanism for retention of anomeric configuration has been proposed involving direct front-side nucleophilic displacement (S<sub>N</sub>i). Both of these mechanisms bear considerable analogy to those proposed for retaining glycosyltransferases <cite>Lairsson2004 Lairsson2008 </cite>. |
[[Image:Retaining_a-glycoside_phosphorylase_mechanism.png|center|600px]] | [[Image:Retaining_a-glycoside_phosphorylase_mechanism.png|center|600px]] | ||
Revision as of 02:07, 20 June 2013
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: Spencer Williams
- Responsible Curator: Spencer Williams
Overview
Phosphorylases are enzymes that catalyze the cleavage of a glycosidic bond through substitution with phosphate (formally referred to as phosphorolysis). Phosphorylases are usually named using a combination of the ‘substrate name’ and ‘phosphorylase’ [1, 2]
Phosphorolysis of a glycosidic bond can occur with retention or inversion of configuration and always occurs in an exo-fashion leading to formation of a monosaccharide or disaccharide 1-phosphate.
The energy content of the sugar 1-phosphate product means that the cleavage reaction is often in a practical sense reversible and, in Nature, these enzymes may be involved in either synthesis or cleavage of the glycosidic bond. As such there is a relatively fine distinction among sugar phosphorylases, glycoside hydrolases and classical sugar nucleoside (di)phosphate dependent glycosyltransferases. In the last case the synthetic reaction is normally, but not always, irreversible because of the higher energy of a sugar nucleoside (di)phosphate.
The phosphorylases are classified into various glycoside hydrolase (GH) and glycosyltransferase (GT) families on the basis of sequence similarity [2].
Glycosyltransferase-like phosphorylases
The classical example of phosphorylases are the glycogen/starch phosphorylases [3]. These enzymes catalyze the cleavage of individual glucosyl residues from glycogen/amylopectin (up to five residues (or up to four for hyperthermophilic bacterial and archael forms) from a branchpoint), forming sequentially deglycosylated glycogen/amylopectin and glucose 1-phosphate. Glycogen/starch phosphorylases have a complex mechanism that is not fully understood and requires pyridoxal phosphate (PLP) as a cofactor. All glycogen/starch phosphorylases are classified into the same sequence-related glycosyltransferase family as starch phosphorylases (GT35), which also require a PLP cofactor. Trehalose phosphorylase (retaining) is classified as a glycosyltransferase and belongs to GT4. There is no evidence that trehalose phosphorylase (retaining) uses a PLP cofactor.
Glycoside hydrolase-like phosphorylases
Most sugar phosphorylases act on glucosides and many cleave simple disaccharides such as sucrose, trehalose, cellobiose and maltose leading to glucose-1-phosphate and the reducing end sugar (glucose or fructose in these specific cases). Other sugar phosphorylases are known that act on N,N'-diacetylchitobiose, laminaribiose, 1,3-β-glucan and nucleosides. A phosphorylase from GH13 has been reported that acts on α-1,4-glucans to release the disaccharide maltose 1-phosphate [4].
Examples
- Family GH13 contains sucrose phosphorylase and α-1,4-glucan:maltose 1-phosphate maltosyltransferase.
- Family GH65 contains maltose phosphorylase, trehalose phosphorylase, kojibiose (Glc-α-1,2-Glc) phosphorylase, nigerose (Glc-α-1,3-Glc) phosphorylase, and trehalose 6-phosphate phosphorylase.
- Family GH94 contains cellobiose phosphorylase, cellodextrin phosphorylase, and chitobiose phosphorylase.
- Family GH112 contains β-galactoside phosphorylases such as β-1,3-D-galactosyl-D-hexososamine phosphorylase, and β-1,4-D-galactosyl-L-rhamnose phosphorylase.
- Family GH130 contains β-mannoside phosphorylases such as β-mannosyl-1,4-glucose phosphorylase and Normal 0 false false false EN-US JA X-NONE and β-1,4-mannooligosaccharide phosphorylase.
Mechanism of glycoside hydrolase-like phosphorylases
Sequence and structural analysis of sugar phosphorylases reveal that some have sequences and structures (and likely mechanisms) similar to glycosyltransferases, whereas others have sequences and structures that more closely resemble glycoside hydrolases.
Inverting phosphorylase mechanism
All inverting phosphorylases are GH-like in their sequence classification. Phosphorolysis of a glycoside by a GH-like phosphorylase with net inversion of anomeric configuration is achieved via a one step, single-displacement mechanism involving oxocarbenium ion-like transition states, as shown below. The reaction occurs with acid/base assistance from a suitably positioned carboxylate residue. This mechanism has clear parallels with the mechanism for inverting glycoside hydrolases.
Retaining phosphorylase mechanism
Phosphorolysis of a glycoside with net retention of configuration by the GH-like retaining phosphorylases is most probably achieved via a two step, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate, as shown in the figure below. Each step passes through an oxocarbenium ion-like transition state. Reaction occurs with acid/base and nucleophilic assistance provided by two amino acid side chains, typically glutamate or aspartate. In the first step (called the glycosylation step), one residue plays the role of a nucleophile, attacking the anomeric centre to displace the aglycon and form a glycosyl enzyme intermediate. At the same time the other residue functions as an acid catalyst and protonates the glycosidic oxygen as the bond cleaves. In the second step (known as the deglycosylation step), the glycosyl enzyme is hydrolyzed by phosphate, with the other residue now acting as a base catalyst deprotonating the phosphate as it attacks. An alternative mechanism for retention of anomeric configuration has been proposed involving direct front-side nucleophilic displacement (SNi). Both of these mechanisms bear considerable analogy to those proposed for retaining glycosyltransferases [3, 5].
References
-
Kitaoka M, Hayashi K: Carbohydrate-processing phosphorolytic enzymes. Trends Glycosci. Glycotechnol. 2002, 14, 35-50.
- Nakai H, Kitaoka M, Svensson B, and Ohtsubo K. (2013). Recent development of phosphorylases possessing large potential for oligosaccharide synthesis. Curr Opin Chem Biol. 2013;17(2):301-9. DOI:10.1016/j.cbpa.2013.01.006 |
- Lairson LL, Henrissat B, Davies GJ, and Withers SG. (2008). Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521-55. DOI:10.1146/annurev.biochem.76.061005.092322 |
- Elbein AD, Pastuszak I, Tackett AJ, Wilson T, and Pan YT. (2010). Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J Biol Chem. 2010;285(13):9803-9812. DOI:10.1074/jbc.M109.033944 |
- Lairson LL and Withers SG. (2004). Mechanistic analogies amongst carbohydrate modifying enzymes. Chem Commun (Camb). 2004(20):2243-8. DOI:10.1039/b406490a |