CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 16
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: Maria Matard-Mann
- Responsible Curator: ^^^Elizabeth Ficko-Blean^^^
| CAZy DB link | |
| https://www.cazy.org/CBM16.html |
Ligand specificities
Family 16 CBMs (CAZy - CBM16) are found essentially in bacteria (with the exception of some CBM16 members in archaea). They are also found associated with catalytic modules belonging mainly to 4 families of CAZymes: GH5 mannanases [1, 2], GH16 kappa carrageenases [3, 4, 5], GH18 chitinases [6] and PL18 alginate lyases [7, 8]. Binding to glucomannan and kappa-carrageenan has been demonstrated [1, 2, 5]. CBM16 binding to glucomannan (mixed β-1,4-linked polymer contains both glucose and mannose) has been studied by mean of ITC (isothermal titration calorimetry) analysis and X-ray crystallography of complexes with pentomannan and pentoglucan [1, 2]. Conversely, binding to kappa-carrageenan has been shown by a double-blind approach using polysaccharide microarrays [5].
Structural Features
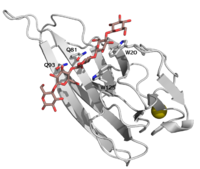
CBM16 is a type B CBM family, with a characteristic concave cleft, allowing the binding of substrate longer than triose. The ligand binding cleft shows some promiscuity as it can accommodate both pentoses (containing glucose and mannose), but only in the context of planar polymer like β-1,4-glucans, and not helical β-1,3-glucans [1]. The crystallographic structure determination of both CBMs from Caldanaerobius polysaccharolyticus (formerly Thermoanaerobacterium polysaccharolyticum) ManA revealed the importance of two aromatic residues in the binding cleft, as long as two stretches of polar residues on both sides of the cleft [1]. Affinity studies of targeted mutants for the predicted key resides confirmed the importance of two tryptophans (Trp-20 and Trp-125), and two glutamines (Gln-81 and Gln-93) [2].
Based on sequence similarity and conservation of secondary structure elements it has been proposed that, along with the CBM4, CBM17, CBM22 and CBM27 families, they form a superfamily [9].
Functionalities
In the Man5A of Caldanaerobius polysaccharolyticus, the deletion of both its CBM16s severely impairs the ability of the catalytic module (GH5) to bind cellulose [10].
In the case of CgkA from Zobellia galactanivorans, the presence of the CBM16 is not required for the enzymatic activity on kappa-carrageenan, but has been shown to take part in the processive mechanism of the catalytic module (GH16) [4].
Even if frequently found within the gene coding for alginate lyase from family PL18, it is absent in the mature form of the enzyme, and no role in alginate degradation has been found up to now [8]. A chaperone function of this N-terminal module has been proposed after observation that its deletion hindered the correct folding and activity of the catalytic module [7].
Family Firsts
- First Identified
Cloning of Man5A GH5 by Cann et al. in 1999 reaveled the presence of two CBM16 tandem associated in C-terminal side. Their deletion resulted in failure of the catalytic module to bind to cellulose column, and significant loss of both mannanase and carboxy methylcellulase activities [10].
- First Structural Characterization
Bae et al. solved in 2008 the first structures of CBM16 family: both modules of Caldanaerobius polysaccharolyticus Man5A, (PDBID: 2zew, 2zez), and two complexes of CBM16-1, one with cellopentaose (PDBID: 2zex) and one with mannopentaose (PDBID: 2zey)[1].
References
- Bae B, Ohene-Adjei S, Kocherginskaya S, Mackie RI, Spies MA, Cann IK, and Nair SK. (2008). Molecular basis for the selectivity and specificity of ligand recognition by the family 16 carbohydrate-binding modules from Thermoanaerobacterium polysaccharolyticum ManA. J Biol Chem. 2008;283(18):12415-25. DOI:10.1074/jbc.M706513200 |
- Su X, Agarwal V, Dodd D, Bae B, Mackie RI, Nair SK, and Cann IK. (2010). Mutational insights into the roles of amino acid residues in ligand binding for two closely related family 16 carbohydrate binding modules. J Biol Chem. 2010;285(45):34665-76. DOI:10.1074/jbc.M110.168302 |
- Barbeyron T, Gerard A, Potin P, Henrissat B, and Kloareg B. (1998). The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol Biol Evol. 1998;15(5):528-37. DOI:10.1093/oxfordjournals.molbev.a025952 |
- Matard-Mann M, Bernard T, Leroux C, Barbeyron T, Larocque R, Préchoux A, Jeudy A, Jam M, Nyvall Collén P, Michel G, and Czjzek M. (2017). Structural insights into marine carbohydrate degradation by family GH16 κ-carrageenases. J Biol Chem. 2017;292(48):19919-19934. DOI:10.1074/jbc.M117.808279 |
- Salmeán AA, Guillouzo A, Duffieux D, Jam M, Matard-Mann M, Larocque R, Pedersen HL, Michel G, Czjzek M, Willats WGT, and Hervé C. (2018). Double blind microarray-based polysaccharide profiling enables parallel identification of uncharacterized polysaccharides and carbohydrate-binding proteins with unknown specificities. Sci Rep. 2018;8(1):2500. DOI:10.1038/s41598-018-20605-9 |
- Barabote RD, Xie G, Leu DH, Normand P, Necsulea A, Daubin V, Médigue C, Adney WS, Xu XC, Lapidus A, Parales RE, Detter C, Pujic P, Bruce D, Lavire C, Challacombe JF, Brettin TS, and Berry AM. (2009). Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations. Genome Res. 2009;19(6):1033-43. DOI:10.1101/gr.084848.108 |
- Dong S, Wei TD, Chen XL, Li CY, Wang P, Xie BB, Qin QL, Zhang XY, Pang XH, Zhou BC, and Zhang YZ. (2014). Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18. J Biol Chem. 2014;289(43):29558-69. DOI:10.1074/jbc.M114.584573 |
- Sim PF, Furusawa G, and Teh AH. (2017). Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2. Sci Rep. 2017;7(1):13656. DOI:10.1038/s41598-017-13288-1 |
- Sunna A, Gibbs MD, and Bergquist PL. (2001). Identification of novel beta-mannan- and beta-glucan-binding modules: evidence for a superfamily of carbohydrate-binding modules. Biochem J. 2001;356(Pt 3):791-8. DOI:10.1042/0264-6021:3560791 |
- Cann IK, Kocherginskaya S, King MR, White BA, and Mackie RI. (1999). Molecular cloning, sequencing, and expression of a novel multidomain mannanase gene from Thermoanaerobacterium polysaccharolyticum. J Bacteriol. 1999;181(5):1643-51. DOI:10.1128/JB.181.5.1643-1651.1999 |