CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 70
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| CAZy DB link | |
| https://www.cazy.org/CBM70.html |
Ligand specificities
The CBM70 family comprises members predominantly of bacterial origin. Notably, it is the only known family with a specific binding affinity for hyaluronic acid, a linear glycosaminoglycan composed of repeating disaccharide units of β-1,4-ᴅ-glucuronic acid-β-1,3-N-acetyl-ᴅ-glucosamine. Studies have shown that CBM70 modules typically do not bind to other glycosaminoglycans, such as chondroitin sulfate, dermatan sulfate, or heparin [1, 2].
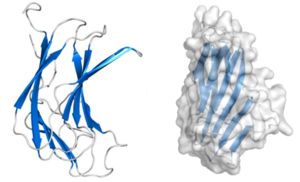
Structural Features
CBM70 modules are typically composed of approximately 160 amino acids. The crystal structure of the N-terminal CBM70 module (SpCBM70) from S. pneumoniae hyaluronate lyase Hyl has been determined. SpCBM70 adopts a classic β-jelly roll fold, consisting of two opposing 5-stranded antiparallel β-sheets. This slightly bowed sandwich structure creates a groove along the concave surface, which carries a significant positive charge and is highly conserved within the CBM70 family [1].
Functionalities
CBM70 domains are commonly found as accessory modules in hyaluronate lyases produced by bacteria of the Streptococcus genus. These domains enhance the enzyme capability to degrade hyaluronic acid, a crucial component of the host's extracellular matrix. During infection, pathogens such as S. pneumoniae utilize hyaluronate lyase to break down hyaluronic acid, facilitating bacterial invasion and spread. CBM70 domains boost this process by increasing the binding efficiency of the enzyme, playing a key role in pathogen virulence and contributing to the high specificity of the enzyme for hyaluronic acid. Additionally, CBM70 domains have been effectively utilized in lateral flow immunoassays for the specific detection of hyaluronic acid, demonstrating their potential in diagnostic applications [2].
Family Firsts
- First Identified
The first CBM70 module to be identified (SpCBM70) was from the S. pneumoniae hyaluronate lyase Hyl.
- First Structural Characterization
The first crystal structure of a CBM70 module was also that of SpCBM70, PDB ID 4D0Q.
References
- Suits MDL, Pluvinage B, Law A, Liu Y, Palma AS, Chai W, Feizi T, and Boraston AB. (2014). Conformational analysis of the Streptococcus pneumoniae hyaluronate lyase and characterization of its hyaluronan-specific carbohydrate-binding module. J Biol Chem. 2014;289(39):27264-27277. DOI:10.1074/jbc.M114.578435 |
- Mei X, Sun M, Zhang Y, Shen J, Li J, Xue C, and Chang Y. (2022). Establishment of a carbohydrate binding module-based lateral flow immunoassay method for identifying hyaluronic acid. Int J Biol Macromol. 2022;223(Pt A):1180-1185. DOI:10.1016/j.ijbiomac.2022.11.122 |