CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 84
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Ian Greig^^^
- Responsible Curator: ^^^David Vocadlo^^^
| Glycoside Hydrolase Family GH84 | |
| Clan | none |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH84.html | |
Substrate specificities
GH84 contains β-N-acetylglucosaminidases and β-N-acetylhyaluronidase activities. Human O-GlcNAcase is a nucleocytoplasmic enzyme whose in vivo targets are glycoprotein serine and threonine residues modified by a single β-linked GlcNAc residue. In contrast to the β-hexosaminidases of GH20 a relaxed specificity for substitutions of the N-acyl group is observed with residues significantly more bulky than the N-acyl group being tolerated.[1]
Kinetics and Mechanism
Members of GH84 utilize a mechanism of neighbouring group participation, this originally being established through the use of free-energy relationship-based studies of human O-GlNAcase.[1] More recent studies of this enzyme have investigated variations in rates of reaction (V/K) with both nucleophile and leaving group structures.[2] For substrates possessing the naturally-occurring acetyl nucleophile a pre-chemical step is rate-determining on V/K for leaving groups with a pKa below 11 (with the chemical step rate-determining for substrates with higher pKa values). Studies of substrates possessing fluoroacetyl nucleophiles highlighted that a dissociative (DN*AN) mechanism involving general acid catalysis operates for the hydrolysis of substrates possessing leaving groups with a pKa greater than approximately 7; a concerted (ANDN) mechanism, not employing general acid catalysis was found for substrates possessing leaving groups with lower pKas (consistent with prior studies of S-glucosaminide hydrolysis[3]).
Numerous carbohydrate and carbohydrate like-scaffolds have been reported as yield potent inhibitors of GH84 enzymes. These include "NAG-thiazolines",REF, PUGNAc (O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate),[4] GlcNAcstatin, [5, 6, 6, 7, 8, 9, 10, 10, 10, 10, 11, 11, 12, 12, 13, 14, 15, 16, 17, 18, 19, 20, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 32, 33, 34, 35, 36, 37, 38, 39, 40, 40, 41, 42, 43, 44, 45, 46, 47, 48] Many of these families of inhibitors possess structural characteristics reminiscent of the oxacarbenium ion-like transition states of glycosyl group transfer and, as such may loosely be termed 'transition state analogues'. An analysis of NAG-thiazoline- and PUGNAc-derived inhibitors of human O-GlcNAcase has shown that only the NAG-thiazolines position the inhibitors and their N-acyl side-chains within the hydrophobic binding pocket in a manner consistent with the species found along the reaction coordinate.[49] As such NAG-thiazoline inhibitors may be termed 'Bartlett-type' (free-energy relationship-based)[50] transition state analogues. Substrate distortion.[2, 51]
A truncated, nuclear-localized isoform of human O-GlcNAcase lacking the putative C-terminal histone acetyl transferase domain retains similar kinetic properties and inhibitory patterns as the full-length cytosolic isoform and is consistent with hexosaminidase activity residing in the N-terminal domain.[52]
Catalytic Residues
Studies of two mutants of human O-GlcNAcase established that adjacent aspartate residues, Asp174 and Asp175, act as critical components of the catalytic machinery of this enzyme.[53]
The mutant Asp175Ala displayed marked reductions in activity (V and (V/K)) towards aryl N-acetylglucosaminides possessing poor leaving groups with smaller reductions being observed for both O-aryl and S-aryl N-acetylglucosaminides substrates possessing better leaving groups. Exogenous azide was found to partially rescue the activity of human O-GlcNAcase towards 3,4-dinitrophenylglucosaminide. These results identify Asp175 as the general acid catalyst.
The mutant Asp174Ala showed decreased activity towards O-aryl N-acetylglucosaminides possessing good leaving groups and it was argued that this is consistent with its role as a residue responsible for the orientation and polarization of the N-acyl nucleophile.
Three-dimensional structures
The reported crystallization of Clostridium perfringens NagJ[54] was followed by solved structures for that enzyme[55] and Bacteroides thetaiotaomicron β-hexosaminidase[55]. In common with the chitinases of family GH18 and the exo-acting β-hexosaminidases of GH20 the catalytic domain is a (βα)8-barrel structure. The structure of human O-GlcNAcase has not been solved but Bacteroides thetaiotaomicron β-hexosaminidase was originally reported as a good structural mimic of both the active site and catalytic domain of human O-GlcNAcase. More recently the structure of the GH84 β-hexosaminidase from Oceanicola granulosus has been solved and shown to possess an improved sequence identity with the catalytic domain of the human enzyme.[56] Furthermore, the C-terminal domain of this protein also displays notable sequence identity with the spacer domain (separating the domains possessing O-GlcNAcase and histone acetyltransferase activities) of the human enzyme.
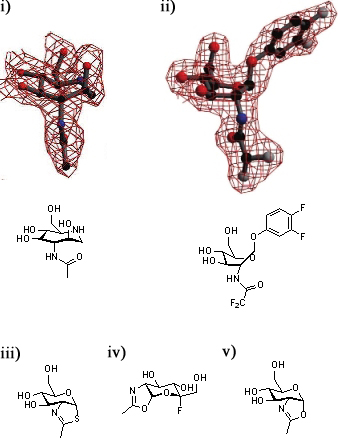
A series of crystallographic studies on Bacteroides thetaiotaomicron β-hexosaminidase have used a variety of small molecules to define define the conformational itinerary for this family of enzymes. Substrate distortion from the 4C1 conformation found in solution to a bound 1,4B / 1S3 conformation was supported by the crystal structure of the wild-type enzyme in complex with the 7-membered azepane. [57] This distortion was confirmed by the structure the wild-type enzyme in complex with the substrate, 3,4-difluorophenyl 2-deoxy-2-difluoroacetamido-β-D-glucopyranoside.[58] Earlier studies of the wild-type-bound thiazoline show that this intermediate is found in a 4C1conformation.[1] Subsequent studies have shown that oxazoline intermediates are bound to the mutant enzymes in this conformation.[58].
Family Firsts
- First sterochemistry determination
- 1H-NMR studies of human O-GlcNAcase established that the β-configured hemiacetal product is formed prior to anomerisation.[2].
- First catalytic nucleophile identification
- This family of enzymes uses a mechanism of neighbouring group participation, which was first establishes through the use of free energy relationships studies.[1].
- First general acid/base residue identification
- Studies of human O-GlcNAcase mutant Asp175Ala identify reactivity patterns (free energy relationships, pH-activity profiles) consistent with the action of Asp175 as the catalytic general acid/base.[53].
- First 3-D structure
- The structures of Bacteroides thetaiotaomicron O-GlcNAcase[59] and Clostridium perfringens NagJ[55].
References
- Macauley MS, Whitworth GE, Debowski AW, Chin D, and Vocadlo DJ. (2005). O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280(27):25313-22. DOI:10.1074/jbc.M413819200 |
- Greig IR, Macauley MS, Williams IH, and Vocadlo DJ. (2009). Probing synergy between two catalytic strategies in the glycoside hydrolase O-GlcNAcase using multiple linear free energy relationships. J Am Chem Soc. 2009;131(37):13415-22. DOI:10.1021/ja904506u |
- Macauley MS, Stubbs KA, and Vocadlo DJ. (2005). O-GlcNAcase catalyzes cleavage of thioglycosides without general acid catalysis. J Am Chem Soc. 2005;127(49):17202-3. DOI:10.1021/ja0567687 |
- Stubbs KA, Zhang N, and Vocadlo DJ. (2006). A divergent synthesis of 2-acyl derivatives of PUGNAc yields selective inhibitors of O-GlcNAcase. Org Biomol Chem. 2006;4(5):839-45. DOI:10.1039/b516273d |
- Dorfmueller HC, Borodkin VS, Schimpl M, Shepherd SM, Shpiro NA, and van Aalten DM. (2006). GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J Am Chem Soc. 2006;128(51):16484-5. DOI:10.1021/ja066743n |
- Dorfmueller HC, Borodkin VS, Schimpl M, Zheng X, Kime R, Read KD, and van Aalten DM. (2010). Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases. Chem Biol. 2010;17(11):1250-5. DOI:10.1016/j.chembiol.2010.09.014 |
- Whitworth GE, Macauley MS, Stubbs KA, Dennis RJ, Taylor EJ, Davies GJ, Greig IR, and Vocadlo DJ. (2007). Analysis of PUGNAc and NAG-thiazoline as transition state analogues for human O-GlcNAcase: mechanistic and structural insights into inhibitor selectivity and transition state poise. J Am Chem Soc. 2007;129(3):635-44. DOI:10.1021/ja065697o |
- Mader MM and Bartlett PA. (1997). Binding Energy and Catalysis: The Implications for Transition-State Analogs and Catalytic Antibodies. Chem Rev. 1997;97(5):1281-1302. DOI:10.1021/cr960435y |
- He Y, Macauley MS, Stubbs KA, Vocadlo DJ, and Davies GJ. (2010). Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J Am Chem Soc. 2010;132(6):1807-9. DOI:10.1021/ja9086769 |
- Macauley MS and Vocadlo DJ. (2009). Enzymatic characterization and inhibition of the nuclear variant of human O-GlcNAcase. Carbohydr Res. 2009;344(9):1079-84. DOI:10.1016/j.carres.2009.04.017 |
-
Cetinbaş N, Macauley MS, Stubbs KA, Drapala R, Vocadlo DJ. Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry. 2006 Mar 21;45(11):3835-44.
Note: Due to a problem with PubMed data, this reference is not automatically formatted. Please see these links out: DOI:10.1021/bi052370b PMID:16533067
- Ficko-Blean E and Boraston AB. (2005). Cloning, recombinant production, crystallization and preliminary X-ray diffraction studies of a family 84 glycoside hydrolase from Clostridium perfringens. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61(Pt 9):834-6. DOI:10.1107/S1744309105024012 |
- Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, and van Aalten DM. (2006). Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25(7):1569-78. DOI:10.1038/sj.emboj.7601026 |
- Schimpl M, Schüttelkopf AW, Borodkin VS, and van Aalten DM. (2010). Human OGA binds substrates in a conserved peptide recognition groove. Biochem J. 2010;432(1):1-7. DOI:10.1042/BJ20101338 |
- Marcelo F, He Y, Yuzwa SA, Nieto L, Jiménez-Barbero J, Sollogoub M, Vocadlo DJ, Davies GD, and Blériot Y. (2009). Molecular basis for inhibition of GH84 glycoside hydrolases by substituted azepanes: conformational flexibility enables probing of substrate distortion. J Am Chem Soc. 2009;131(15):5390-2. DOI:10.1021/ja809776r |
- He Y, Macauley MS, Stubbs KA, Vocadlo DJ, and Davies GJ. (2010). Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J Am Chem Soc. 2010;132(6):1807-9. DOI:10.1021/ja9086769 |
- Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, and Davies GJ. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol. 2006;13(4):365-71. DOI:10.1038/nsmb1079 |
- Comfort DA, Bobrov KS, Ivanen DR, Shabalin KA, Harris JM, Kulminskaya AA, Brumer H, and Kelly RM. (2007). Biochemical analysis of Thermotoga maritima GH36 alpha-galactosidase (TmGalA) confirms the mechanistic commonality of clan GH-D glycoside hydrolases. Biochemistry. 2007;46(11):3319-30. DOI:10.1021/bi061521n |
- He S and Withers SG. (1997). Assignment of sweet almond beta-glucosidase as a family 1 glycosidase and identification of its active site nucleophile. J Biol Chem. 1997;272(40):24864-7. DOI:10.1074/jbc.272.40.24864 |
-
Sinnott, M.L. (1990) Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev. 90, 1171-1202. DOI: 10.1021/cr00105a006