CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 102
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM102.html |
Ligand specificities
The dissected C-terminal CBM102IV of a surface glycan-binding protein (SGBP; containing four CBM102s I-IV, Figure 1a) of Christiangramia forsetii KT0803T binds laminarin (from Laminaria digitata) and laminarin-derived oligosaccharides as determined by affinity gel electrophoresis and/or isothermal titration calorimetry [1]. CBM102IV has a higher affinity to laminarin (Ka ~1.04 x 105 M-1) than to laminariheptaose (Ka ~1.87 x 104 M-1) and laminaripentaose (Ka ~1.39 x 104 M-1). Laminaribiose did not bind the CBM102IV. In addition, the dissected C-terminal CBM102IV has a higher affinity to laminarin than two CBM102 tandem constructs of the SGBP (Ka ~2.76 x 104 M-1 or ~3.63 x 104 M-1) and the full length SGBP (Ka ~3.09 x 104 M-1) [1].
Structural Features
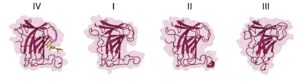
The CBM102 displays a β-sandwich fold established by two antiparallel β-sheets forming a convex and a concave face. The 3D X-ray crystal structure of the C-terminal CBM102IV of the C. forsetii SGBP showed that, together with two expansive loops, the concave face provides a narrow cleft to accommodate laminaritriose [1] (Figure 1b). CBM102 thus represents a type B CBM. Three residues located in the concave face mediate polar interactions with laminaritriose (Lys825, Glu827 and Glu837), while one aromatic residue of each loop (Tyr790, Phe861) is involved in CH-π interaction with the ligand (Figure 1c). However, the C. forsetii protein contains three additional CBM102s. Alphafold2 [2] predictions showed that two of them are highly similar to the 3D crystal structure of the C-terminal CBM102IV, but that one CBM102 is missing one of the loops shaping the pocket, which results in a very shallow cleft (Figure 2). This was speculated to represent an adaptation to highly branched laminarins [1].

Functionalities
The CBM102-containing SGBP is tethered to the outer membrane of C. forsetii and is abundantly produced in laminarin-grown cells [3]. With four CBM102s, the SGBP provides multiple laminarin docking sites [1]. The protein further contains two N-terminal Ig-like domains that precede the CBM102s, which presumably act as spacers to ensure distance to the membrane and exposure of binding sites as suggested before for β-glucan-binding CBM-containing SGBPs [4]. The four repeating CBM102s in the C. forsetii protein were suggested to render the SGBP an optimal 'sugar-trapping antenna' on the bacterial surface; this is supported by an elongated protein shape (Rg 52 Å and Dmax 178 Å) as determined by small angle X-ray scattering [1]. Unexpectedly, isothermal titration calorimetry showed that maximum two laminarin molecules were bound by the four CBM102s and that affinity was not increased by multiple CBM102s. In bacterial metagenome-assembled genomes from phytoplankton blooms in the North Sea, CBM102 was detected together with appended catalytic modules, indicating other putative functions (for example enhancing enzyme concentration on substrate to improve catalysis) [1]. The majority of associated catalytic modules were GH16_3s, but also rare examples of GH2, GH5_34, GH81, and GH162. Within this dataset, CBM102 also co-occurred with CBM4, CBM6 and CBM103 in multimodular proteins. CBM102-only-proteins contained up to eight repeats of CBM102. The production of CBM102-containing proteins correlated with the course of the phytoplankton bloom, which underlines the relevance of CBM102 in marine β-glucan use.
Family Firsts
- First Identified
- CBM102 was first identified in an SGBP encoded in a laminarin utilization locus of Christiangramia forsetii KT0803T [1], a marine flavobacterium isolated from surface waters in the North Sea [5]. More than 350 CBM102-containing sequences were detected in bacterial metagenome-assembled genomes (555 representative MAGs of which 201 belonged to Bacteroidota) retrieved from bacterial biomass from phytoplankton blooms of three respective years (2016, 2018 and 2020) in the North Sea. Sequences mostly belonged to Bacteroidota, but also to Proteobacteria, Myxococcota and Actinobacteriota [1]. The association with GHs (mostly GH16_3) within this dataset led to the creation of the CBM family 102.
- First Structural Characterization
- The C-terminal CBM102IV of the laminarin PUL-encoded SGBP of C. forsetii KT0803T is the first structurally characterized member of the family (PDB ID 8QX6). The 3D crystal structure was obtained in complex with laminaritriose.
References
Error fetching PMID 35637307:
Error fetching PMID 24522261:
Error fetching PMID 33587952:
Error fetching PMID 11679337:
- Error fetching PMID 38757353:
- Error fetching PMID 35637307:
- Error fetching PMID 24522261:
- Error fetching PMID 33587952:
- Error fetching PMID 11679337: