CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycosyltransferase Family 138
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycosyltransferase Family GT138 | |
| Clan | Fido fold |
| Mechanism | Inverting |
| Active site residues | Known |
| CAZy DB link | |
| https://www.cazy.org/GT138.html | |
Substrate specificities
The GT138 family of glycosyltransferases is exemplified by AvrB [1]. As a bacterial effector from the plant pathogen Pseudomonas syringae, AvrB utilizes host UDP-rhamnose (or dTDP-rhamnose in vitro) as a co-substrate to rhamnosylate the host protein RIN4, and causes the programmed cell death (i.e. the hypersensitive response) [1, 2]. AvrB contains a "Fido" domain [3, 4], different from other known glycosyltransferases (see below). Interestingly, Fido proteins can also have AMPylation [5], phosphorylation [6], UMPylation [7], and phosphocholination [8, 9] activities. Hence, AvrB is a unique Fido protein that functions as a glycosyltransferase.
Kinetics and Mechanism
In the reaction, rhamnose is directly transferred to the side chain of a threonine of RIN4, T166 (Fig. 1) [1]. The rhamnosylation reaction catalyzed by AvrB does not require divalent cations (e.g., Mg2+) [1].
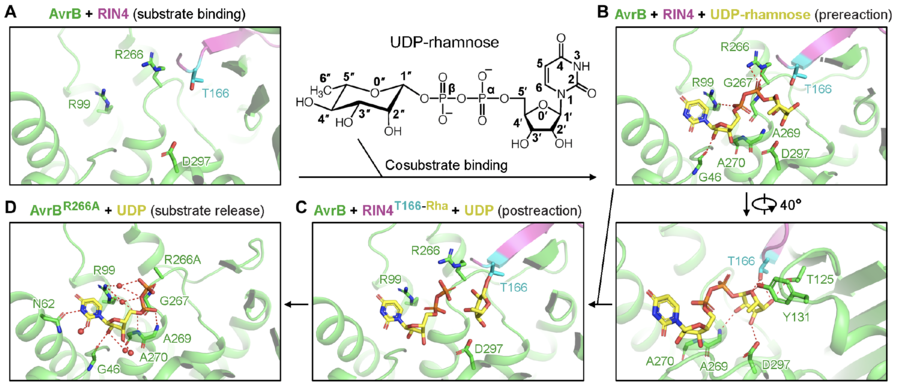
Catalytic Residues
A threonine (T166) from the protein substrate directly attacks the rhamnose moiety in the co-substrate, UDP-rhamnose (Fig. 1) [1]. The threonine is close to a histidine and a threonine in AvrB, which may stabilize the acceptor. UDP-rhamnose is stabilized by a few residues in the pocket (Fig. 1) [1].
Three-dimensional structures
AvrB represents the prototype for glycosyltransferases comprised of a Fido fold (Fig. 2A) [1]. Most glycosyltransferases contain GT-A, GT-B, GT-C, lysozyme-type, GT101, or GT108 folds (Fig. 2B) [10, 11, 12, 13]. AvrB contains a large internal domain between helix α2 and helix α3 (Fig. 2A) [1, 3, 4, 14]. AvrB shares similar structural features with other Fido proteins despite substantial primary sequence divergence [4].
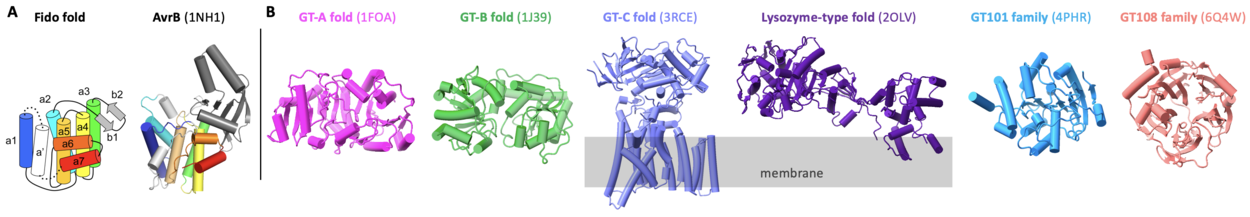
Family Firsts
The first member of GT138 family shown to be a glycosyltransferase is AvrB [1].
The first structure of GT138 family is AvrB (Fig. 2A) [3]. A few AvrB structures are available to reveal the catalysis mechanisms [1, 3, 14]
References
- Peng W, Garcia N, Servage KA, Kohler JJ, Ready JM, Tomchick DR, Fernandez J, and Orth K. (2024). Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4. Sci Adv. 2024;10(7):eadd5108. DOI:10.1126/sciadv.add5108 |
- Mackey D, Holt BF 3rd, Wiig A, and Dangl JL. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108(6):743-54. DOI:10.1016/s0092-8674(02)00661-x |
- Lee CC, Wood MD, Ng K, Andersen CB, Liu Y, Luginbühl P, Spraggon G, and Katagiri F. (2004). Crystal structure of the type III effector AvrB from Pseudomonas syringae. Structure. 2004;12(3):487-94. DOI:10.1016/j.str.2004.02.013 |
- Kinch LN, Yarbrough ML, Orth K, and Grishin NV. (2009). Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One. 2009;4(6):e5818. DOI:10.1371/journal.pone.0005818 |
- Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, and Orth K. (2009). AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323(5911):269-72. DOI:10.1126/science.1166382 |
- Castro-Roa D, Garcia-Pino A, De Gieter S, van Nuland NAJ, Loris R, and Zenkin N. (2013). The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol. 2013;9(12):811-7. DOI:10.1038/nchembio.1364 |
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, and Zhou JM. (2012). A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485(7396):114-8. DOI:10.1038/nature10962 |
- Mukherjee S, Liu X, Arasaki K, McDonough J, Galán JE, and Roy CR. (2011). Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477(7362):103-6. DOI:10.1038/nature10335 |
- Campanacci V, Mukherjee S, Roy CR, and Cherfils J. (2013). Structure of the Legionella effector AnkX reveals the mechanism of phosphocholine transfer by the FIC domain. EMBO J. 2013;32(10):1469-77. DOI:10.1038/emboj.2013.82 |
- Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, and Seeberger PH. (2022). 2022. DOI:10.1101/9781621824213 |
- Lairson LL, Henrissat B, Davies GJ, and Withers SG. (2008). Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521-55. DOI:10.1146/annurev.biochem.76.061005.092322 |
- Zhang H, Zhu F, Yang T, Ding L, Zhou M, Li J, Haslam SM, Dell A, Erlandsen H, and Wu H. (2014). The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat Commun. 2014;5:4339. DOI:10.1038/ncomms5339 |
- Sernee MF, Ralton JE, Nero TL, Sobala LF, Kloehn J, Vieira-Lara MA, Cobbold SA, Stanton L, Pires DEV, Hanssen E, Males A, Ward T, Bastidas LM, van der Peet PL, Parker MW, Ascher DB, Williams SJ, Davies GJ, and McConville MJ. (2019). A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites. Cell Host Microbe. 2019;26(3):385-399.e9. DOI:10.1016/j.chom.2019.08.009 |
- Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, and Dangl JL. (2007). Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 2007;3(3):e48. DOI:10.1371/journal.ppat.0030048 |