CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycosyltransferase Family 37
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: Vivek Bharadwaj, Hsin-Tzu Wang, and Breeanna Urbanowicz
- Responsible Curator: Breeanna Urbanowicz
| Glycosyltransferase Family GT37 | |
| Fold | GT-B |
| Mechanism | Inverting |
| Active site residues | Known |
| CAZy DB link | |
| https://www.cazy.org/GT37.html | |
Substrate specificities
The enzymes belonging to the GT37 family are Golgi localized, plant-specific fucosyltransferases (FUTs) that utilize guanidine 5’-diphosphate-β-L-Fucose (GDP-Fuc) as a donor substrate, and catalyze the transfer of fucose to saccharide acceptor substrates. There are 10 members of this family in the model plant species Arabidopsis thaliana (AtFUT1-10), seven members in Populus trichocarpa (PtFUT1-7). The known acceptor substrates for GT37 FUTs are cell wall glycans, including xyloglucans (XyGs) and arabinogalactan proteins (AGPs). It is hypothesized that members of this family may also be involved in the synthesis of the pectic polysaccharides rhamnogalacturonan I (RG-I) and rhamnogalacturonan II (RG-II). All biochemically characterized GT37 FUTs to date are α-(1-2)-fucosyltransferases [1].
XyGs are hemicellulosic polysaccharides composed of a β-(1,4)-linked glucosyl backbone with a variety of sidechains initiated with an α-D-xylose (Xyl) at O-6 position. The xylosyl residue on the sidechain can be further substituted with β-D-galactose (Gal), β-D-galacturonic acid (GalA), α-L- Arabinofuranose (Araf), or α-L-Arabinopyranose (Arap)depending on the species and tissue [2]. Gal, GalA, and Arap be further substituted by an α-L-fucose (Fuc) through an α-(1-2) linkage [3, 4]. So far, there are three FUTs that have been identified and characterized to be specifically involved in fucosylating the Gal or GalA in XyGs, including PsFUT1 [5], AtFUT1 [6] and OsMUR2 [7, 8].
FUTs are also involved in AGPs synthesis. AGPs are a diverse family of cell wall glycoproteins with large arabinogalactan polysaccharide chains attached to non-contiguous hydroxyproline residues. The backbone of the O-glycan chains on AGPs is composed of β-1,3-linked galactosyl residues that are branched by β-1,6-linked galactosyl sidechains which are further substituted by α-L-Araf and β-D-(methyl)glucuronic acid (GlcA). Araf and GlcA on the sidechains can then be decorated by α-L-Fuc and α-L-Rhamnose (Rha), respectively, as well as other sugars such as α-D-Xyl [9]. Three of the family members in GT37, AtFUT4, AtFUT6 and AtFUT7 from A. thaliana, are involved in fucosylating AGPs [10, 11]. A previous study done using tobacco Bright Yellow-2 (BY-2) suspension cells indicated that the AtFUT4- and AtFUT6-overexpressed BY-2 cells produced AGPs with Fuc attached to the O-2 position of α-L-Ara [11]. Recently, a glycan array-based assay using recombinant AtFUT4, AtFUT6 and AtFUT7 showed that all three enzymes have a broad substrate specificity and are able to fucosylate oligosaccharides containing Araf residues α-1,3-linked to Gal, structures representative of AGPs, as well as β-1,4-xylotertrose appended with an α-1,3-linked Araf at the non-reducing end [10].
There is an α-L-Fuc α-(1-2)-linked to β-D-Gal on in RG-I, and a 2-O-Me-α-L-Fuc that is attached to β-D-Gal via an α-(1-2)-linkage on sidechain B of RG-II, it is hypothesized that the uncharacterized members of the GT37 family are involved in the synthesis of these structures [1, 12].
Kinetics and Mechanism
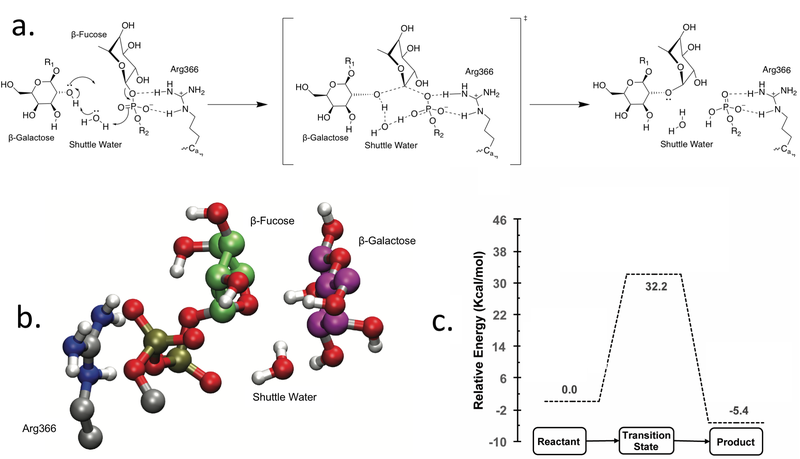
GT-37 enzymes are classified as inverting GTs i.e. the anomeric configuration of the transferred sugar is inverted in the final product Coutinho2003. The recent structural characterization of AtFUT1 provided detailed insights into the kinetics and mechanism of one GT-37 enzyme [13, 14] In general, inverting GTs are considered to employ a direct displacement SN2-like reaction catalyzed by a catalytic general base at the active site to deprotonate the acceptor oxygen for nucleophilic attack [15]. However, sequence, structural and biochemical analysis revealed no identifiable catalytic base residue at the active site in AtFUT1 [13]. Based on molecular dynamics simulations and quantum mechanical calculations, a water mediated semi-concerted SN1-like reaction mechanism, consistent with biochemical mutagenesis data was demonstrated for AtFUT1. The water facilitates catalysis as a proton shuttle that initially receives a proton from the acceptor galactose residue (O2 hydroxyl) to generate the nucleophile, which attacks the fucose residue on the donor. Simultaneously, the shuttle water donates its other proton to phosphate of the nucleotide, which acts as the base to complete the reaction mechanism [13].
Catalytic Residues
An arginine residue (Arg366 in AtFUT1) is established to be indispensable for catalysis, as it binds the nucleotide-sugar at the active site and stabilizes the oxygen atoms on the phosphate group [13]. In the reaction mechanism, this arginine residue facilitates the release of the fucose group by stabilizing the developing negative charge on the phosphate oxygens, consistent with its indispensable role in catalysis. In light of the absence of a single proximal catalytic general base in AtFUT1, mutagenic and computational studies reveal that multiple residues are observed to play a critical role in AtFUT1 catalysis. A H-bonding network involving Asp550, Tyr486 and His523 is proposed to be crucial for activating the shuttle water molecule to initiate catalysis and stabilizing the transition state [13].
Three-dimensional structures
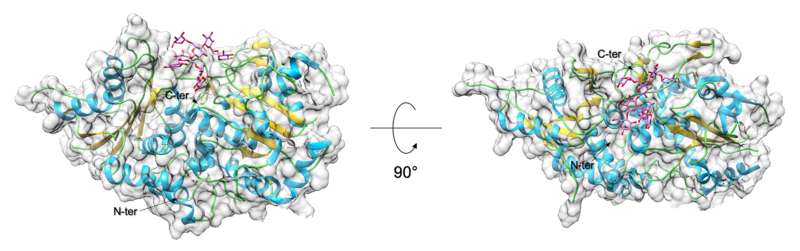
GT structures have in general been classified as either belonging to GT-A or GT-B fold [16]. Amongst the GT-37 family of enzymes, only AtFUT1 has been structurally characterized [13, 14]. The AtFUT1 structure is observed to adopt a GT-B type fold, with two b/a/b Rossmann-like domains. The metal independent catalytic site is located in between the two Rossmann domains and binds the nucleotide-sugar donor. A solvent exposed substrate binding groove is created at the interface between these two domains and binds the galactose containing acceptor oligosaccharide.
Family Firsts
- First 3-D structure
- The AtFUT1 enzyme was the first GT-37 family to be structurally characterized using X-ray Crystallography. 5 structures of AtFUT1 (apo PDB ID 5KOP [14], XXLG bound PDB ID 5KOE [13], GDP bound PDB ID 5KWK [13], and XXLG-GDP complex PDB ID 5KOR [14]) are available in PDB.
References
-
Maria J. Soto, Breeanna R. Urbanowicz and Michael G. Hahn (2019) Plant Fucosyltransferases and the Emerging Biological Importance of Fucosylated Plant Structures. Critical Reviews in Plant Sciences 38, 327-338 DOI:10.1080/07352689
- Peña MJ, Kong Y, York WS, and O'Neill MA. (2012). A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell. 2012;24(11):4511-24. DOI:10.1105/tpc.112.103390 |
-
Fry, S. C., York, W. S., Albersheim, P., Darvill, A., Hayashi, T., Joseleau, J.-P., Kato, Y., Lorences, E. P., Maclachlan, G. A., McNeil, M., Mort, A. J., Grant Reid, J. S., Seitz, H. U., Selvendran, R. R., Voragen, A. G. J., and White, A. R. 1993. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89: 1–3. DOI:10.1111/j.1399-3054.1993.tb01778.x
- Tuomivaara ST, Yaoi K, O'Neill MA, and York WS. (2015). Generation and structural validation of a library of diverse xyloglucan-derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydr Res. 2015;402:56-66. DOI:10.1016/j.carres.2014.06.031 |
- Farkas V and Maclachlan G. (1988). Fucosylation of exogenous xyloglucans by pea microsomal membranes. Arch Biochem Biophys. 1988;264(1):48-53. DOI:10.1016/0003-9861(88)90568-1 |
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, and Keegstra K. (1999). Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284(5422):1976-9. DOI:10.1126/science.284.5422.1976 |
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, and Reiter WD. (2002). The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci U S A. 2002;99(5):3340-5. DOI:10.1073/pnas.052450699 |
- Liu L, Paulitz J, and Pauly M. (2015). The presence of fucogalactoxyloglucan and its synthesis in rice indicates conserved functional importance in plants. Plant Physiol. 2015;168(2):549-60. DOI:10.1104/pp.15.00441 |
- Showalter AM and Basu D. (2016). Extensin and Arabinogalactan-Protein Biosynthesis: Glycosyltransferases, Research Challenges, and Biosensors. Front Plant Sci. 2016;7:814. DOI:10.3389/fpls.2016.00814 |
- Ruprecht C, Bartetzko MP, Senf D, Lakhina A, Smith PJ, Soto MJ, Oh H, Yang JY, Chapla D, Varon Silva D, Clausen MH, Hahn MG, Moremen KW, Urbanowicz BR, and Pfrengle F. (2020). A Glycan Array-Based Assay for the Identification and Characterization of Plant Glycosyltransferases. Angew Chem Int Ed Engl. 2020;59(30):12493-12498. DOI:10.1002/anie.202003105 |
- Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, and Faik A. (2010). Functional identification of two nonredundant Arabidopsis alpha(1,2)fucosyltransferases specific to arabinogalactan proteins. J Biol Chem. 2010;285(18):13638-45. DOI:10.1074/jbc.M110.102715 |
- Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, Keegstra K, and Raikhel NV. (2001). Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol. 2001;127(4):1595-606. | Google Books | Open Library
- Urbanowicz BR, Bharadwaj VS, Alahuhta M, Peña MJ, Lunin VV, Bomble YJ, Wang S, Yang JY, Tuomivaara ST, Himmel ME, Moremen KW, York WS, and Crowley MF. (2017). Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism. Plant J. 2017;91(6):931-949. DOI:10.1111/tpj.13628 |
- Rocha J, Cicéron F, de Sanctis D, Lelimousin M, Chazalet V, Lerouxel O, and Breton C. (2016). Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation. Plant Cell. 2016;28(10):2352-2364. DOI:10.1105/tpc.16.00519 |
- Lairson LL, Henrissat B, Davies GJ, and Withers SG. (2008). Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521-55. DOI:10.1146/annurev.biochem.76.061005.092322 |
- Breton C, Fournel-Gigleux S, and Palcic MM. (2012). Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol. 2012;22(5):540-9. DOI:10.1016/j.sbi.2012.06.007 |
- Coutinho PM, Deleury E, Davies GJ, and Henrissat B. (2003). An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328(2):307-17. DOI:10.1016/s0022-2836(03)00307-3 |