CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 164"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (24 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Zachary Armstrong|Zachary Armstrong]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Gideon Davies|Gideon Davies]] |
---- | ---- | ||
| Line 12: | Line 12: | ||
|- | |- | ||
|'''Clan''' | |'''Clan''' | ||
| − | |GH- | + | |GH-A |
|- | |- | ||
|'''Mechanism''' | |'''Mechanism''' | ||
| − | |retaining | + | |retaining |
|- | |- | ||
|'''Active site residues''' | |'''Active site residues''' | ||
| − | | | + | |known |
|- | |- | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
| Line 26: | Line 26: | ||
</div> | </div> | ||
<!-- This is the end of the table --> | <!-- This is the end of the table --> | ||
| − | |||
== Substrate specificities == | == Substrate specificities == | ||
| − | + | [[Image:BS164_AB.png|thumb|right|300px|'''Figure 1. '''The trimeric structure of Bs164 is shown in panel''' A'''. All three protomers are shown with a surface and each chain is displayed as a cartoon diagram coloured by domain.''' B '''shows the structure of one protomer. Domain A, which has a (β/α)8 fold, is shown in green with subdomain H is shown in magenta, domain B, containing a mixed β-sheet, is shown in red and the β-sandwich of domain C is shown in blue. ''See also PDB IDs [{{PDBlink}}6T5O 6T5O], [{{PDBlink}}6T75 6T75], [{{PDBlink}}6T7G 6T7G], and [{{PDBlink}}6T6G 6T6G].'']] | |
| − | + | The defining member of [[glycoside hydrolase]] family 164, a β-mannosidase from ''Bacteroides salyersiae'' (''Bs''164, GenbankID: EIY59668.1), was identified initially identified through rational bioinformatic selection of enzyme targets <cite>Helbert2019</cite>. Although ''Bs''164 was initially reported as an α-mannosidase, subsequent detailed biochemical characterization and structure determination revealed that was instead a β-mannosidase <cite>Armstrong2020</cite>. This enzyme is an exo-acting and is capable of cleaving mannooligos and β-mannosides<cite>Armstrong2020</cite>. | |
| − | |||
| − | |||
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | + | ''Bs''164 β-mannosidase is a [[retaining]] enzyme, as first shown by NMR <cite>Armstrong2020</cite>, and follows the [[classical Koshland double-displacement mechanism]]. | |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | + | The catalytic nucleophile of ''Bs''164 was identified as glutamate 297 through mutational analysis<cite>Armstrong2020</cite>. A structural complex with 2,4-dinitrophenyl 2-deoxy-2-fluoro-β-D-mannopyranoside showed a covalent attachment of the inhibitor to glutamate 297, reinforcing the assignment of Glu297 as the catalytic nucleophile. The acid/base residue, Glu160 is positioned to perform [[Syn/anti lateral protonation|''anti''-protonation]] of the leaving group, typical of clan GH-A glycoside hydrolases. This residue forms hydrogen bonding interactions with both the endocyclic nitrogen in noeuromycin and imidazole nitrogen in mannoimidazole. Complete loss of activity by the E160Q variant confirmed the assignment of Glu160 as the acid/base residue<cite>Armstrong2020</cite>. | |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | + | The structure of ''Bs''164 was solved using multi-wavelength anomalous diffraction of a seleno-methionine labeled protein <cite>Armstrong2020</cite>. The structure of ''Bs''164 has been solved in an uncomplexed state ([{{PDBlink}}6T5O PDB ID 6T5O]), in complex with mannoimidazole ([{{PDBlink}}6T7G PDB ID 6T7G]) and noeuromycin ([{{PDBlink}}6T6G PDB ID 6T6G]), and as a covalent 2-deoxy-2-fluoromannosyl-enzyme intermediate ([{{PDBlink}}6T75 PDB ID 6T75]). Bs164 exists as a donut shaped trimer (Figure 1A). Each trimer-donut has an outer diameter of approximately 100 Å and an internal diameter of between 30 and 35 Å. The individual Bs164 chains contain three clearly defined domains: a modified (β/α)<sub>8</sub> barrel, a domain containing a seven membered mixed β-sheet sandwiched between α-helices, and a β-sheet domain (Figure 1B). The catalytic residues are present on strands 4 (acid/base) and 7 (nucleophile) of the (β/α)<sub>8</sub> barrel indicating that GH164 belongs to clan GH-A glycoside hydrolases. This domain architecture is quite similar to that seen for family [[GH42]] enzymes <cite>Hidaka2002</cite>, but is previously unseen for β-mannosidases. | |
| − | |||
== Family Firsts == | == Family Firsts == | ||
;First sterochemistry determination: ''Bacteroides salyersiae'' β-mannosidase by NMR <cite>Armstrong2020</cite> | ;First sterochemistry determination: ''Bacteroides salyersiae'' β-mannosidase by NMR <cite>Armstrong2020</cite> | ||
| Line 52: | Line 48: | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Helbert2019 pmid=30850540 |
| − | # | + | #Hidaka2002 pmid=12215416 |
| + | #Armstrong2020 pmid=31871050 | ||
| + | |||
</biblio> | </biblio> | ||
[[Category:Glycoside Hydrolase Families|GH164]] | [[Category:Glycoside Hydrolase Families|GH164]] | ||
| − | |||
Latest revision as of 13:19, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH164 | |
| Clan | GH-A |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH164.html | |
Substrate specificities
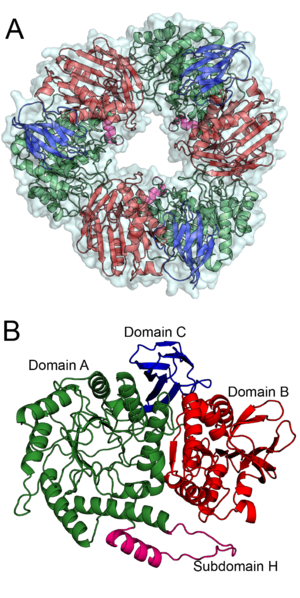
The defining member of glycoside hydrolase family 164, a β-mannosidase from Bacteroides salyersiae (Bs164, GenbankID: EIY59668.1), was identified initially identified through rational bioinformatic selection of enzyme targets [1]. Although Bs164 was initially reported as an α-mannosidase, subsequent detailed biochemical characterization and structure determination revealed that was instead a β-mannosidase [2]. This enzyme is an exo-acting and is capable of cleaving mannooligos and β-mannosides[2].
Kinetics and Mechanism
Bs164 β-mannosidase is a retaining enzyme, as first shown by NMR [2], and follows the classical Koshland double-displacement mechanism.
Catalytic Residues
The catalytic nucleophile of Bs164 was identified as glutamate 297 through mutational analysis[2]. A structural complex with 2,4-dinitrophenyl 2-deoxy-2-fluoro-β-D-mannopyranoside showed a covalent attachment of the inhibitor to glutamate 297, reinforcing the assignment of Glu297 as the catalytic nucleophile. The acid/base residue, Glu160 is positioned to perform anti-protonation of the leaving group, typical of clan GH-A glycoside hydrolases. This residue forms hydrogen bonding interactions with both the endocyclic nitrogen in noeuromycin and imidazole nitrogen in mannoimidazole. Complete loss of activity by the E160Q variant confirmed the assignment of Glu160 as the acid/base residue[2].
Three-dimensional structures
The structure of Bs164 was solved using multi-wavelength anomalous diffraction of a seleno-methionine labeled protein [2]. The structure of Bs164 has been solved in an uncomplexed state (PDB ID 6T5O), in complex with mannoimidazole (PDB ID 6T7G) and noeuromycin (PDB ID 6T6G), and as a covalent 2-deoxy-2-fluoromannosyl-enzyme intermediate (PDB ID 6T75). Bs164 exists as a donut shaped trimer (Figure 1A). Each trimer-donut has an outer diameter of approximately 100 Å and an internal diameter of between 30 and 35 Å. The individual Bs164 chains contain three clearly defined domains: a modified (β/α)8 barrel, a domain containing a seven membered mixed β-sheet sandwiched between α-helices, and a β-sheet domain (Figure 1B). The catalytic residues are present on strands 4 (acid/base) and 7 (nucleophile) of the (β/α)8 barrel indicating that GH164 belongs to clan GH-A glycoside hydrolases. This domain architecture is quite similar to that seen for family GH42 enzymes [3], but is previously unseen for β-mannosidases.
Family Firsts
- First sterochemistry determination
- Bacteroides salyersiae β-mannosidase by NMR [2]
- First catalytic nucleophile identification
- Bacteroides salyersiae β-mannosidase by 2-fluoromannose labeling and kinetic analysis of mutants [2]
- First general acid/base residue identification
- Bacteroides salyersiae β-mannosidase by kinetic analysis of mutants [2]
- First 3-D structure of a GH1 enzyme
- Bacteroides salyersiae β-mannosidase [2]
References
- Helbert W, Poulet L, Drouillard S, Mathieu S, Loiodice M, Couturier M, Lombard V, Terrapon N, Turchetto J, Vincentelli R, and Henrissat B. (2019). Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc Natl Acad Sci U S A. 2019;116(13):6063-6068. DOI:10.1073/pnas.1815791116 |
- Armstrong Z and Davies GJ. (2020). Structure and function of Bs164 β-mannosidase from Bacteroides salyersiae the founding member of glycoside hydrolase family GH164. J Biol Chem. 2020;295(13):4316-4326. DOI:10.1074/jbc.RA119.011591 |
- Hidaka M, Fushinobu S, Ohtsu N, Motoshima H, Matsuzawa H, Shoun H, and Wakagi T. (2002). Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J Mol Biol. 2002;322(1):79-91. DOI:10.1016/s0022-2836(02)00746-5 |