CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 14"
Eva Madland (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (19 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Eva Madland|Eva Madland]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Elizabeth Ficko-Blean|Elizabeth Ficko-Blean]] |
---- | ---- | ||
| Line 12: | Line 12: | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
|- | |- | ||
| − | | colspan="2" |{{CAZyDBlink}} | + | | colspan="2" |{{CAZyDBlink}}CBM14.html |
|} | |} | ||
</div> | </div> | ||
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | Family 14 CBMs are modules composed of approximately 70 residues. These modules have been reported to be associated with chitinases <cite>Fadel2016</cite> and as chitin-binding lectins e.g. | + | Family 14 CBMs are modules composed of approximately 70 residues. These modules have been reported to be associated with chitinases <cite>Fadel2016</cite> and as chitin-binding lectins e.g. effector proteins from the tomato pathogens ''Pseudoercospora fuligena'' or ''Cladosporium fulvum'' <cite>Kohler2016 Hurlburt2018</cite>, as an antimicrobial protein (tachycitin) from horseshoe crab (''Tachypleus tridentatus'') hemocytes <cite>Kawabata1996</cite> and in peritrophic matrix proteins from the malaria vector ''Anopheles gambia'' <cite>Shen1998</cite>. Members of CBM14 have been shown to bind chitin <cite>Shen1998 Vandevenne2011 Madland2019</cite> and chitooligomers <cite>Crasson2016 Hurlburt2018</cite>; binding to 50 % acetylated hyaluronan has also been demonstrated <cite>Crasson2016</cite>. |
| − | The ligand binding affinities have been quantified for two CBM14 members. The interaction between a human chitotriosidase-1 (CHIT1, characterized as a glycoside hydrolase family 18 ([[GH18]])) | + | The ligand binding affinities have been quantified for two CBM14 members. The interaction between a CBM14 from human chitotriosidase-1 (CHIT1, characterized as a glycoside hydrolase family 18 ([[GH18]])) and (GlcNAc)<sub>3</sub> has been investigated. The CBM14 displayed a relatively weak interaction of K<sub>D</sub> 9.9 ± 0.8 mM using NMR titration experiments <cite>Crasson2016</cite> and K<sub>D</sub> 3.1 ± 0.2 mM using isothermal titration calorimetry (ITC) <cite>Madland2019</cite>. |
| − | Interaction studies have also been performed for CBM14 | + | Interaction studies have also been performed for a CBM14 from the fungal tomato pathogen ''C. fulvum'' and (GlcNAc)<sub>6</sub>. Here, the binding properties were measured using ITC yielding a K<sub>D</sub> of 6.7 ± 1.5 µM <cite>Hurlburt2018</cite>. |
== Structural Features == | == Structural Features == | ||
| − | [[File:Fig1-cbm14.png|thumb|300px|right|'''Figure 1 Example of CBM14 structures.''' A) CBM14 | + | [[File:Fig1-cbm14.png|thumb|300px|right|'''Figure 1 Example of CBM14 structures.''' A) ''Hs''CBM14 (PDB ID: [{{PDBlink}}6SO0 6SO0]) cartoon representation. Residues involved in binding (Trp465 and Asn466) are highlighted in cyan, Leu454 in purple, the two aromatic residues (Phe437 and Phe456) forming a hydrophobic core in white and disulfide briges in orange. B) Surface representation of ''Hs''CBM14. C) ''Cf''CBM14 (PDB ID: [{{PDBlink}}6BN0 6BN0]) cartoon representation. Residues involved in binding (Lys49, Cys50, Met51, Pro53, Lys99, Trp100, Cys101, Asp102 and Tyr103) are highlighted in cyan, the two aromatic residues (Tyr76 and Trp92) forming the hydrophobic core in white, disulfide briges in orange and (GlcNAc)<sub>6</sub> in blue. D) Surface representation of ''Cf''CBM14.]] |
| − | + | CBM14 members have a hevein-like fold made up by a central β-sheet (three anti-parallel β-strands) linked to a small β-sheet (two anti-parallel β-strands) by two aromatic residues. The CBM14 members from tomato pathogens also have an N-terminal α-helix and an extended loop connecting -strands 2 and 3, as well as a C-terminal helical turn <cite>Kohler2016 Hurlburt2018</cite>. The latter is also present in tachycitin <cite>Suetake2000</cite>. In addtion, the CBM14 structures have been reported to contain 3-4 disulfide bridges which also serve as a stabilizing effect on this unique fold <cite>Suetake2000 Fadel2016 Hurlburt2018</cite>. | |
| − | With | + | With the hevein-like fold and lectin-like properties, members of the CBM14 family are characterized as [[Carbohydrate-binding_modules#Types|type C]] CBMs <cite>Boraston2004</cite>. This is in line with the ligand-binding feature seen for the CBM14 associated with CHIT1 (''Homo sapiens'' CBM14, ''Hs''CBM14). The binding site is composed of a tryptophan and an asparagine (Trp465 and Asn466) at the C-terminus of the module <cite>Crasson2016</cite> (Figure 1A). These residues create a platform-like binding surface (Figure 1B) commonly seen in [[Carbohydrate-binding_modules#Types|type A]] CBMs and could be the reason for why ''Hs''CBM14 is also able to bind crystalline chitin. In addition to Trp465 and Asn466, a leucine (Leu454) has been reported to be indirectly involved in binding as it contributes to maintaining a correct orientation for Trp465 <cite>Madland2019</cite>. Binding between ''Hs''CBM14 and chitotriose is likely to occur through CH-π stacking between the side-chain of Trp465 and the middle pyranose ring and hydrogen bonds between the side-chain of Asn466 and the hydrophilic non-reducing end <cite>Madland2019</cite>. |
| − | CBM14 from the tomato pathogen ''C. fulvum'' (''Cf''CBM14) was the first of these modules to be co-crystallized with (GlcNAc)<sub>6</sub> (PDB ID: [ | + | CBM14 from the tomato pathogen ''C. fulvum'' (''Cf''CBM14) was the first of these modules to be co-crystallized with (GlcNAc)<sub>6</sub> (PDB ID: [{{PDBlink}}6BN0 6BN0])<cite>Hurlburt2018</cite>. Although the fold of ''Cf''CBM14 is the same as ''Hs''CBM14 (PDB ID: [{{PDBlink}}5HBF 5HBF] and [{{PDBlink}}6SO0 6SO0]) and other [http://www.cazy.org/CBM14_structure.html structurally characterized CBM14 members], the residues responsible for binding differ. Binding of (GlcNAc)<sub>6</sub> is mediated by Lys49, Cys50, Met51, Pro53, Lys99, Trp100, Cys101, Asp102 and Tyr103 positioned along a shallow trench in the longitudinal axis of ''Cf''CBM14 (Figure 1C and D). Mutational studies showed that binding to (GlcNAc)<sub>6</sub> was abolished by mutation of Trp100 and Asp102. The main interaction is reported to be a CH-π stacking between Trp100 and GlcNAc-5 aided by Met51 and Pro53 which are both in van der Waal distances to GlcNAc-1 and GlcNAc-3, respectively <cite>Hurlburt2018</cite>. Although equivalents to Trp100, Asp102 and Tyr103 have been reported to be important for chitin-binding in CBM14 from tomato pathogen ''P. fuligena'' (''Pf''CBM14) (equivalent residues: Trp94, Asp96 and Tyr97) <cite>Kohler2016</cite>, the extensive hydrogen bond network could point to a different binding mechanism for ''Cf''CBM14 that is more reminiscent of a [[Carbohydrate-binding_modules#Types|type B]] CBM. |
== Functionalities == | == Functionalities == | ||
| − | Several members of CBM14 are often encountered as chitin-binding lectins <cite>Kawabata1996 Burg2004 Suetake2000 Kohler2016 Hurlburt2018</cite>, while the members present in modular chitinases play a key role in targeting substrate. The most common associated modules are chitinases, e.g. [[GH18]] from human chitotriosidase structurally determined together with CBM14 | + | Several members of CBM14 are often encountered as chitin-binding lectins <cite>Kawabata1996 Burg2004 Suetake2000 Kohler2016 Hurlburt2018</cite>, while the members present in modular chitinases play a key role in targeting substrate <cite>Renkema1997 Tjoelker2000 Fadel2016</cite>. The most common associated modules are chitinases, e.g. [[GH18]] from human chitotriosidase structurally determined together with ''Hs''CBM14 (PDB ID: [{{PDBlink}}5HBF 5HBF])<cite>Fadel2016</cite>. |
| − | Although some of the structural features described above differ between the | + | Although some of the structural features described above differ between the CBM14 lectins and ''Hs''CBM14, the common denominator is the involvement of these modules in immune responses <cite>Burg2006 Joosten1997 Fadel2016</cite>. CHIT1 is produced by macrophages and neutrophils <cite>Hollak1994, Kzhyshkowska2007</cite> and is utilized by the innate immune response to combat chitin-containing pathogens <cite>Gordon-Thomson2009</cite>. A different response is found in the tomato pathogen ''C. fulvum''; here, the effector protein utilizes CBM14's ability to bind chitin in the fungal cell wall enabling protection from hydrolysis by plant-derived chitinases during infection <cite>Joosten1997 Burg2006</cite>. |
== Family Firsts == | == Family Firsts == | ||
| − | ;First Identified | + | ;First Identified: CBM14 was first identified as an antimicrobial protein with chitin-binding ability in the hemocyte of horseshoe crab (''T. tridentatus'') and called tachycitin <cite>Kawabata1996</cite>. |
| − | :CBM14 was first identified as an antimicrobial | + | ;First Structural Characterization: The first three-dimensional structure was determined by NMR spectroscopy for the CBM14 module tachycitin (PDB ID: [{{PDBlink}}1DQC 1DQC]) <cite>Suetake2000</cite>. The structure of the first carbohydrate-active enzyme associated CBM14 was determined by X-ray crystallography for the CBM14 of human chitotriosidase (PDB ID: [{{PDBlink}}5HBF 5HBF]) <cite>Fadel2016</cite>. |
| − | ;First Structural Characterization | ||
| − | :The first three-dimensional structure was determined by NMR spectroscopy for the CBM14 module tachycitin (PDB ID: [ | ||
== References == | == References == | ||
| Line 53: | Line 51: | ||
#Suetake2000 pmid=10770921 | #Suetake2000 pmid=10770921 | ||
#Boraston2004 pmid=15214846 | #Boraston2004 pmid=15214846 | ||
| + | #Burg2004 pmid=14769793 | ||
| − | # | + | #Renkema1997 pmid=9118991 |
| + | |||
| + | #Tjoelker2000 pmid=10617646 | ||
#Hollak1994 pmid=8132768 | #Hollak1994 pmid=8132768 | ||
#Kzhyshkowska2007 pmid=19662198 | #Kzhyshkowska2007 pmid=19662198 | ||
Latest revision as of 13:20, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM14.html |
Ligand specificities
Family 14 CBMs are modules composed of approximately 70 residues. These modules have been reported to be associated with chitinases [1] and as chitin-binding lectins e.g. effector proteins from the tomato pathogens Pseudoercospora fuligena or Cladosporium fulvum [2, 3], as an antimicrobial protein (tachycitin) from horseshoe crab (Tachypleus tridentatus) hemocytes [4] and in peritrophic matrix proteins from the malaria vector Anopheles gambia [5]. Members of CBM14 have been shown to bind chitin [5, 6, 7] and chitooligomers [3, 8]; binding to 50 % acetylated hyaluronan has also been demonstrated [8].
The ligand binding affinities have been quantified for two CBM14 members. The interaction between a CBM14 from human chitotriosidase-1 (CHIT1, characterized as a glycoside hydrolase family 18 (GH18)) and (GlcNAc)3 has been investigated. The CBM14 displayed a relatively weak interaction of KD 9.9 ± 0.8 mM using NMR titration experiments [8] and KD 3.1 ± 0.2 mM using isothermal titration calorimetry (ITC) [7].
Interaction studies have also been performed for a CBM14 from the fungal tomato pathogen C. fulvum and (GlcNAc)6. Here, the binding properties were measured using ITC yielding a KD of 6.7 ± 1.5 µM [3].
Structural Features
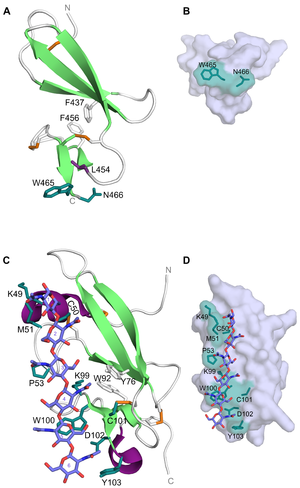
CBM14 members have a hevein-like fold made up by a central β-sheet (three anti-parallel β-strands) linked to a small β-sheet (two anti-parallel β-strands) by two aromatic residues. The CBM14 members from tomato pathogens also have an N-terminal α-helix and an extended loop connecting -strands 2 and 3, as well as a C-terminal helical turn [2, 3]. The latter is also present in tachycitin [9]. In addtion, the CBM14 structures have been reported to contain 3-4 disulfide bridges which also serve as a stabilizing effect on this unique fold [1, 3, 9]. With the hevein-like fold and lectin-like properties, members of the CBM14 family are characterized as type C CBMs [10]. This is in line with the ligand-binding feature seen for the CBM14 associated with CHIT1 (Homo sapiens CBM14, HsCBM14). The binding site is composed of a tryptophan and an asparagine (Trp465 and Asn466) at the C-terminus of the module [8] (Figure 1A). These residues create a platform-like binding surface (Figure 1B) commonly seen in type A CBMs and could be the reason for why HsCBM14 is also able to bind crystalline chitin. In addition to Trp465 and Asn466, a leucine (Leu454) has been reported to be indirectly involved in binding as it contributes to maintaining a correct orientation for Trp465 [7]. Binding between HsCBM14 and chitotriose is likely to occur through CH-π stacking between the side-chain of Trp465 and the middle pyranose ring and hydrogen bonds between the side-chain of Asn466 and the hydrophilic non-reducing end [7].
CBM14 from the tomato pathogen C. fulvum (CfCBM14) was the first of these modules to be co-crystallized with (GlcNAc)6 (PDB ID: 6BN0)[3]. Although the fold of CfCBM14 is the same as HsCBM14 (PDB ID: 5HBF and 6SO0) and other structurally characterized CBM14 members, the residues responsible for binding differ. Binding of (GlcNAc)6 is mediated by Lys49, Cys50, Met51, Pro53, Lys99, Trp100, Cys101, Asp102 and Tyr103 positioned along a shallow trench in the longitudinal axis of CfCBM14 (Figure 1C and D). Mutational studies showed that binding to (GlcNAc)6 was abolished by mutation of Trp100 and Asp102. The main interaction is reported to be a CH-π stacking between Trp100 and GlcNAc-5 aided by Met51 and Pro53 which are both in van der Waal distances to GlcNAc-1 and GlcNAc-3, respectively [3]. Although equivalents to Trp100, Asp102 and Tyr103 have been reported to be important for chitin-binding in CBM14 from tomato pathogen P. fuligena (PfCBM14) (equivalent residues: Trp94, Asp96 and Tyr97) [2], the extensive hydrogen bond network could point to a different binding mechanism for CfCBM14 that is more reminiscent of a type B CBM.
Functionalities
Several members of CBM14 are often encountered as chitin-binding lectins [2, 3, 4, 9, 11], while the members present in modular chitinases play a key role in targeting substrate [1, 12, 13]. The most common associated modules are chitinases, e.g. GH18 from human chitotriosidase structurally determined together with HsCBM14 (PDB ID: 5HBF)[1].
Although some of the structural features described above differ between the CBM14 lectins and HsCBM14, the common denominator is the involvement of these modules in immune responses [1, 14, 15]. CHIT1 is produced by macrophages and neutrophils [16, 17] and is utilized by the innate immune response to combat chitin-containing pathogens [18]. A different response is found in the tomato pathogen C. fulvum; here, the effector protein utilizes CBM14's ability to bind chitin in the fungal cell wall enabling protection from hydrolysis by plant-derived chitinases during infection [14, 15].
Family Firsts
- First Identified
- CBM14 was first identified as an antimicrobial protein with chitin-binding ability in the hemocyte of horseshoe crab (T. tridentatus) and called tachycitin [4].
- First Structural Characterization
- The first three-dimensional structure was determined by NMR spectroscopy for the CBM14 module tachycitin (PDB ID: 1DQC) [9]. The structure of the first carbohydrate-active enzyme associated CBM14 was determined by X-ray crystallography for the CBM14 of human chitotriosidase (PDB ID: 5HBF) [1].
References
- Fadel F, Zhao Y, Cousido-Siah A, Ruiz FX, Mitschler A, and Podjarny A. (2016). X-Ray Crystal Structure of the Full Length Human Chitotriosidase (CHIT1) Reveals Features of Its Chitin Binding Domain. PLoS One. 2016;11(4):e0154190. DOI:10.1371/journal.pone.0154190 |
- Kohler AC, Chen LH, Hurlburt N, Salvucci A, Schwessinger B, Fisher AJ, and Stergiopoulos I. (2016). Structural Analysis of an Avr4 Effector Ortholog Offers Insight into Chitin Binding and Recognition by the Cf-4 Receptor. Plant Cell. 2016;28(8):1945-65. DOI:10.1105/tpc.15.00893 |
- Hurlburt NK, Chen LH, Stergiopoulos I, and Fisher AJ. (2018). Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein. PLoS Pathog. 2018;14(8):e1007263. DOI:10.1371/journal.ppat.1007263 |
- Kawabata S, Nagayama R, Hirata M, Shigenaga T, Agarwala KL, Saito T, Cho J, Nakajima H, Takagi T, and Iwanaga S. (1996). Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J Biochem. 1996;120(6):1253-60. DOI:10.1093/oxfordjournals.jbchem.a021549 |
- Shen Z and Jacobs-Lorena M. (1998). A type I peritrophic matrix protein from the malaria vector Anopheles gambiae binds to chitin. Cloning, expression, and characterization. J Biol Chem. 1998;273(28):17665-70. DOI:10.1074/jbc.273.28.17665 |
- Vandevenne M, Campisi V, Freichels A, Gillard C, Gaspard G, Frère JM, Galleni M, and Filée P. (2011). Comparative functional analysis of the human macrophage chitotriosidase. Protein Sci. 2011;20(8):1451-63. DOI:10.1002/pro.676 |
- Madland E, Crasson O, Vandevenne M, Sørlie M, and Aachmann FL. (2019). NMR and Fluorescence Spectroscopies Reveal the Preorganized Binding Site in Family 14 Carbohydrate-Binding Module from Human Chitotriosidase. ACS Omega. 2019;4(26):21975-21984. DOI:10.1021/acsomega.9b03043 |
- Crasson O, Courtade G, Léonard RR, Aachmann FL, Legrand F, Parente R, Baurain D, Galleni M, Sørlie M, and Vandevenne M. (2017). Human Chitotriosidase: Catalytic Domain or Carbohydrate Binding Module, Who's Leading HCHT's Biological Function. Sci Rep. 2017;7(1):2768. DOI:10.1038/s41598-017-02382-z |
- Suetake T, Tsuda S, Kawabata S, Miura K, Iwanaga S, Hikichi K, Nitta K, and Kawano K. (2000). Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif. J Biol Chem. 2000;275(24):17929-32. DOI:10.1074/jbc.C000184200 |
- Boraston AB, Bolam DN, Gilbert HJ, and Davies GJ. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769-81. DOI:10.1042/BJ20040892 |
- van den Burg HA, Spronk CA, Boeren S, Kennedy MA, Vissers JP, Vuister GW, de Wit PJ, and Vervoort J. (2004). Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein-protein interactions: the chitin-binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin-binding domain. J Biol Chem. 2004;279(16):16786-96. DOI:10.1074/jbc.M312594200 |
- Renkema GH, Boot RG, Strijland A, Donker-Koopman WE, van den Berg M, Muijsers AO, and Aerts JM. (1997). Synthesis, sorting, and processing into distinct isoforms of human macrophage chitotriosidase. Eur J Biochem. 1997;244(2):279-85. DOI:10.1111/j.1432-1033.1997.00279.x |
- Tjoelker LW, Gosting L, Frey S, Hunter CL, Trong HL, Steiner B, Brammer H, and Gray PW. (2000). Structural and functional definition of the human chitinase chitin-binding domain. J Biol Chem. 2000;275(1):514-20. DOI:10.1074/jbc.275.1.514 |
- van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, and de Wit PJ. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact. 2006;19(12):1420-30. DOI:10.1094/MPMI-19-1420 |
- Joosten MH, Vogelsang R, Cozijnsen TJ, Verberne MC, and De Wit PJ. (1997). The biotrophic fungus Cladosporium fulvum circumvents Cf-4-mediated resistance by producing unstable AVR4 elicitors. Plant Cell. 1997;9(3):367-79. DOI:10.1105/tpc.9.3.367 |
- Hollak CE, van Weely S, van Oers MH, and Aerts JM. (1994). Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J Clin Invest. 1994;93(3):1288-92. DOI:10.1172/JCI117084 |
- Kzhyshkowska J, Gratchev A, and Goerdt S. (2007). Human chitinases and chitinase-like proteins as indicators for inflammation and cancer. Biomark Insights. 2007;2:128-46. | Google Books | Open Library
- Gordon-Thomson C, Kumari A, Tomkins L, Holford P, Djordjevic JT, Wright LC, Sorrell TC, and Moore GP. (2009). Chitotriosidase and gene therapy for fungal infections. Cell Mol Life Sci. 2009;66(6):1116-25. DOI:10.1007/s00018-009-8765-7 |