CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 189"
| Line 33: | Line 33: | ||
The reaction products of TiCGS<sub>Cy</sub> were obtained using LβG as the substrate. The β-glucosidase-resistant compounds in the products were purified by size-exclusion chromatography. <sup>1</sup>H-NMR analysis of these purified polysaccharides suggested that they are CβGs. These results clearly demonstrate that TiCGS<sub>Cy</sub> has a retaining mechanism <cite>Tanaka2024</cite>. | The reaction products of TiCGS<sub>Cy</sub> were obtained using LβG as the substrate. The β-glucosidase-resistant compounds in the products were purified by size-exclusion chromatography. <sup>1</sup>H-NMR analysis of these purified polysaccharides suggested that they are CβGs. These results clearly demonstrate that TiCGS<sub>Cy</sub> has a retaining mechanism <cite>Tanaka2024</cite>. | ||
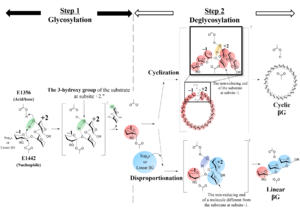
| − | Structural analysis (see | + | Structural analysis (see “Three-dimensional structures” below) and mutational analysis suggest that E1442 of TiCGS<sub>Cy</sub> acts on an anomeric carbon of a glucose moiety at subsite −1 as a nucleophile and E1356 of TiCGS<sub>Cy</sub> acts on a scissile bond of a substrate via 3-hydroxy group of a glucose moiety at subsite +2 as an acid/base <cite>Tanaka2024</cite>. The reaction mechanism of TiCGS<sub>Cy</sub> is noncanonical in that a substrate hydroxy group participates in the catalytic process <cite>Tanaka2024</cite>.[[File:Fig1.png|thumb]] |
== Catalytic Residues == | == Catalytic Residues == | ||
| Line 40: | Line 40: | ||
As of Jan. 2024, the detailed reaction mechanisms of [[GH144]] enzymes remain unclear <cite>Abe2017</cite>. However, SGLs belonging to both [[GH144]] and [[GH162]] have an inverting mechanism unlike [[GH189]] <cite>Tanaka2024,Tanaka2019,Abe2017</cite>. A structural comparison revealed that the positions of the acid/base residue in [[GH189]], the candidate catalytic residues in [[GH144]] and the general acid residue in [[GH162]] are well superimposed. On the other hand, the positions of the other catalytic residues (or candidate catalytic residues) in [[GH189]], [[GH144]] and [[GH162]] are completely different. | As of Jan. 2024, the detailed reaction mechanisms of [[GH144]] enzymes remain unclear <cite>Abe2017</cite>. However, SGLs belonging to both [[GH144]] and [[GH162]] have an inverting mechanism unlike [[GH189]] <cite>Tanaka2024,Tanaka2019,Abe2017</cite>. A structural comparison revealed that the positions of the acid/base residue in [[GH189]], the candidate catalytic residues in [[GH144]] and the general acid residue in [[GH162]] are well superimposed. On the other hand, the positions of the other catalytic residues (or candidate catalytic residues) in [[GH189]], [[GH144]] and [[GH162]] are completely different. | ||
| − | == | + | == Three-dimensional structure == |
The apo-structure of the recombinant TiCGS<sub>Cy</sub> was determined at 3.8 Å by X-ray crystal structure analysis (PDB: 8WY1) <cite>Tanaka2024</cite>. The overall structure comprises a single (α/α)<sub>6</sub>-barrel domain with several inserted α-helices and TiCGS<sub>Cy</sub> has a large active-site pocket <cite>Tanaka2024</cite>. Interestingly, although [[GH189]] (= TiCGS<sub>Cy</sub>), [[GH144]] and [[GH162]] enzymes are quite different in their amino acid sequences, their overall structures and the positions of the substrate in their catalytic pockets are similar <cite>Tanaka2024</cite>. | The apo-structure of the recombinant TiCGS<sub>Cy</sub> was determined at 3.8 Å by X-ray crystal structure analysis (PDB: 8WY1) <cite>Tanaka2024</cite>. The overall structure comprises a single (α/α)<sub>6</sub>-barrel domain with several inserted α-helices and TiCGS<sub>Cy</sub> has a large active-site pocket <cite>Tanaka2024</cite>. Interestingly, although [[GH189]] (= TiCGS<sub>Cy</sub>), [[GH144]] and [[GH162]] enzymes are quite different in their amino acid sequences, their overall structures and the positions of the substrate in their catalytic pockets are similar <cite>Tanaka2024</cite>. | ||
| Line 49: | Line 49: | ||
;First catalytic nucleophile identification: The structural and mutational analysis of TiCGS<sub>Cy</sub> <cite>Tanaka2024</cite>. | ;First catalytic nucleophile identification: The structural and mutational analysis of TiCGS<sub>Cy</sub> <cite>Tanaka2024</cite>. | ||
;First general acid/base residue identification: The structural and mutational analysis of TiCGS<sub>Cy</sub> <cite>Tanaka2024</cite>. | ;First general acid/base residue identification: The structural and mutational analysis of TiCGS<sub>Cy</sub> <cite>Tanaka2024</cite>. | ||
| − | ;First | + | ;First three-dimensional structure: The apo-structure of the recombinant TiCGS<sub>Cy</sub> was determined by X-ray crystal structure analysis <cite>Tanaka2024</cite>. |
== References == | == References == | ||
Revision as of 23:04, 1 February 2024
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Authors: Tomoko Masaike, Masahiro Nakajima, and Nobukiyo Tanaka
- Responsible Curator: Masahiro Nakajima
| Glycoside Hydrolase Family GH189 | |
| Clan | GH-x |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH189.html | |
Substrate specificities
The cyclization domain alone of cyclic β-1,2-glucan synthase from Thermoanaerobacter italicus (TiCGSCy) was identified, characterized and structurally analyzed as reported in 2024 [1]. This enzyme established the novel glycoside hydrolase family (GH) 189. This enzyme specifically catalyzes transglycosylation reactions on linear β-1,2-glucans (LβGs) and β-1,2-glucooligosaccharides (Sopns, where 'n' represents the degree of polymerization (DP)) with DP 6 or more [1]. In the deglycosylation step, intermolecular transglycosylation results in release of disproportionated linear products, while intramolecular transglycosylation results in cyclization of the substrates to release cyclic β-1,2-glucans (CβGs) [1].
Kinetics and Mechanism
The reaction products of TiCGSCy were obtained using LβG as the substrate. The β-glucosidase-resistant compounds in the products were purified by size-exclusion chromatography. 1H-NMR analysis of these purified polysaccharides suggested that they are CβGs. These results clearly demonstrate that TiCGSCy has a retaining mechanism [1].
Structural analysis (see “Three-dimensional structures” below) and mutational analysis suggest that E1442 of TiCGSCy acts on an anomeric carbon of a glucose moiety at subsite −1 as a nucleophile and E1356 of TiCGSCy acts on a scissile bond of a substrate via 3-hydroxy group of a glucose moiety at subsite +2 as an acid/base [1]. The reaction mechanism of TiCGSCy is noncanonical in that a substrate hydroxy group participates in the catalytic process [1].
Catalytic Residues
The acid/base residue and the nucleophile residue of TiCGSCy are E1356 and E1442, respectively. In addition, these residues are well superimposed with the general acid and the nucleophilic water in the GH162 fungal β-1,2-glucanase from Talaromyces funiculosus (TfSGL), respectively [1, 2]. TfSGL has an inverting mechanism particularly catalyzing a unique reaction via the 3-hydroxy group of the glucose molecule at subsite +2 [2].
As of Jan. 2024, the detailed reaction mechanisms of GH144 enzymes remain unclear [3]. However, SGLs belonging to both GH144 and GH162 have an inverting mechanism unlike GH189 [1, 2, 3]. A structural comparison revealed that the positions of the acid/base residue in GH189, the candidate catalytic residues in GH144 and the general acid residue in GH162 are well superimposed. On the other hand, the positions of the other catalytic residues (or candidate catalytic residues) in GH189, GH144 and GH162 are completely different.
Three-dimensional structure
The apo-structure of the recombinant TiCGSCy was determined at 3.8 Å by X-ray crystal structure analysis (PDB: 8WY1) [1]. The overall structure comprises a single (α/α)6-barrel domain with several inserted α-helices and TiCGSCy has a large active-site pocket [1]. Interestingly, although GH189 (= TiCGSCy), GH144 and GH162 enzymes are quite different in their amino acid sequences, their overall structures and the positions of the substrate in their catalytic pockets are similar [1].
GH144 and GH162 were officially classified as part of clan GH-S in this paper [1]. Although GH189 enzymes share a similar (α/α)6 fold structure with GH144 and GH162 enzymes, GH189 enzymes have a retaining mechanism unlike GH144 and GH162 enzymes with an inverting mechanism. Due to the fundamental difference in reaction mechanisms, GH189 is not included in clan GH-S; instead, GH189 could be regarded as a member of a potential new GH clan [1].
Family Firsts
- First stereochemistry determination
- 1H-NMR analysis of β-glucosidase-resistant compounds, which produced by TiCGSCy using LβG as a substrate, as described above [1].
- First catalytic nucleophile identification
- The structural and mutational analysis of TiCGSCy [1].
- First general acid/base residue identification
- The structural and mutational analysis of TiCGSCy [1].
- First three-dimensional structure
- The apo-structure of the recombinant TiCGSCy was determined by X-ray crystal structure analysis [1].
References
- Tanaka N, Saito R, Kobayashi K, Nakai H, Kamo S, Kuramochi K, Taguchi H, Nakajima M, and Masaike T. (2024). Functional and structural analysis of a cyclization domain in a cyclic β-1,2-glucan synthase. Appl Microbiol Biotechnol. 2024;108(1):187. DOI:10.1007/s00253-024-13013-9 |
- Tanaka N, Nakajima M, Narukawa-Nara M, Matsunaga H, Kamisuki S, Aramasa H, Takahashi Y, Sugimoto N, Abe K, Terada T, Miyanaga A, Yamashita T, Sugawara F, Kamakura T, Komba S, Nakai H, and Taguchi H. (2019). Identification, characterization, and structural analyses of a fungal endo-β-1,2-glucanase reveal a new glycoside hydrolase family. J Biol Chem. 2019;294(19):7942-7965. DOI:10.1074/jbc.RA118.007087 |
- Abe K, Nakajima M, Yamashita T, Matsunaga H, Kamisuki S, Nihira T, Takahashi Y, Sugimoto N, Miyanaga A, Nakai H, Arakawa T, Fushinobu S, and Taguchi H. (2017). Biochemical and structural analyses of a bacterial endo-β-1,2-glucanase reveal a new glycoside hydrolase family. J Biol Chem. 2017;292(18):7487-7506. DOI:10.1074/jbc.M116.762724 |