CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycosyltransferase Family 1
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| Glycosyltransferase Family GT1 | |
| Fold | GT-B, 2 Rosmann domains |
| Mechanism | Inverting via a SN2 (O-, N-, S-) or SEAr (C-) |
| Active site residues | Generally a His/Asp dyad as catalytic base |
| CAZy DB link | |
| https://www.cazy.org/GT1.html | |
Substrate specificities
Glycosyl donor: nucleotide sugar. Family 1 glycosyltransferases or GT1s [1, 2, 3] are more often refered to as UGTs, for UDP-dependent glycosyltransferases. Indeed, the most common glycosyl donnor substrates are UDP-α-D-glycosyl, but other nucleotide sugars can be found as well. They are thus also part of the Leloir glycosyltransferases. To date (July 2024), there are no described CAZy subfamilies of GT1s. On the other hand, over hundred subfamilies of GT1s are described in the UGT naming convention system, which organise the enzymes in funtion of the kingdom of their provenance (plant, animal, bacteria) and by phylogeny, based on a system developped for UDP-glucuronyltransferases [4]. This convention is widely used to name the individual enzymes as well, allowing from the name of the enzyme to know which subfamily it belongs to.
Glycosyl acceptor: very diverse. The glycosyl acceptors of GT1 enzymes or UGTs are particularly diverse. In several organisms, from humans to planta to insects, one of their major biological role is the metabolization and detoxification of harmful chemicals. Hence they are largely promiscuous, and a single organism can display over 100 different ones. Efforts have been made to characterize UGTs acceptor specificity at large scale, as well as to predict it [5, 6]. As acceptor substrate specificity is loosely connected to phylogeny, the enzymes names using the UGT nomenclature system also hints about which class of acceptors are most probable for a given enzyme.
Kinetics and Mechanism
The overall mechanism of GT1 enzymes features a catalytic base which activates the glycosyl acceptor, while the nucleotide glycosyl donor dissociate into an ion-pair between an oxocarbenium-like glycosyl and the phosphate of the nucleotide [7]. The mechanism is metal ion-independent. Classically, GT1s have kcat in the order of magnitude of 1 per second, corresponding to an energy barrier of 17 kcal/mol, and KM in the order of the tens of µM. GT1s very often present an acceptor substrate inhibition, which can intriguingly be alleviated with higher enzymes concentrations [8].
Catalytic Residues
The large majority of GT1 presents a His-Asp catalytic dyad [9], acting as a general base. The histidine abstract a proton, increasing the nucleophilicity of the glycosyl acceptor. The aspartate activates the histidine, the abstracted proton being shared almost equally between these two residues [7]. However, this His-Asp dyad is not completely conserved, and among the earliest GT1 studied, GtfA, GtfB [10], and GtfD present an aspartate as a base.
Three-dimensional structures
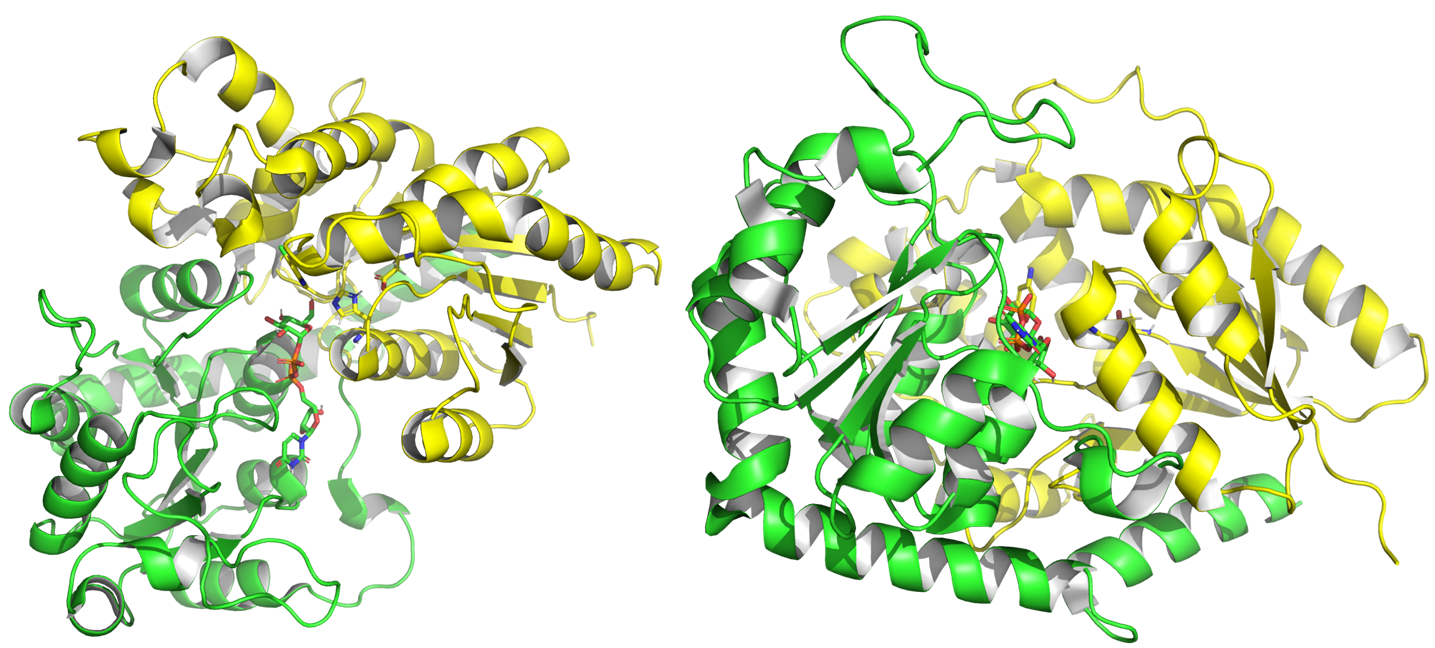
From 2001 to July 2024, experimental structures of 75 different GT1 enzymes have been deposited into the PDB, including with donors and acceptors. GT1 present a GT-B fold [11], characterized by two Rossman domains. The N-terminal domain binds the glycosyl acceptor site (+1), and the C-terminal one (-1) binds the glycosyl donor, usually a UDP alpha glycosyl.
Structure of PtUGT1 from Polygonum tinctorum (PDB ID 6SU6). N-terminal Rossmann domain is represented in yellow, the C-terminal one in green. The two substrates and two catalytic residues are represented with sticks.
Family Firsts
The GT1 family was first proposed in 1997 [1, 12].
First 3D structure, GtfB (Amycolatopsis orientalis) PDB ID 1IIR in 2001 [10].
First His-Asp as catalytic dyad (His20 Asp119) [9]. First Asp as catalytic base [10].
References
- Campbell JA, Davies GJ, Bulone V, and Henrissat B. (1997). A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326 ( Pt 3)(Pt 3):929-39. DOI:10.1042/bj3260929u |
-
Davies, G.J. and Sinnott, M.L. (2008) Sorting the diverse: the sequence-based classifications of carbohydrate-active enzymes. The Biochemist, vol. 30, no. 4., pp. 26-32. DOI:10.1042/BIO03004026.
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, and Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233-8. DOI:10.1093/nar/gkn663 |
- Ross J, Li Y, Lim E, and Bowles DJ. (2001). Higher plant glycosyltransferases. Genome Biol. 2001;2(2):REVIEWS3004. DOI:10.1186/gb-2001-2-2-reviews3004 |
- Yang M, Fehl C, Lees KV, Lim EK, Offen WA, Davies GJ, Bowles DJ, Davidson MG, Roberts SJ, and Davis BG. (2018). Functional and informatics analysis enables glycosyltransferase activity prediction. Nat Chem Biol. 2018;14(12):1109-1117. DOI:10.1038/s41589-018-0154-9 |
- Harding-Larsen D, Madsen CD, Teze D, Kittilä T, Langhorn MR, Gharabli H, Hobusch M, Otalvaro FM, Kırtel O, Bidart GN, Mazurenko S, Travnik E, and Welner DH. (2024). GASP: A Pan-Specific Predictor of Family 1 Glycosyltransferase Acceptor Specificity Enabled by a Pipeline for Substrate Feature Generation and Large-Scale Experimental Screening. ACS Omega. 2024;9(25):27278-27288. DOI:10.1021/acsomega.4c01583 |
-
Teze, D.; Coines, J.; Fredslund, F.; Dubey, K. D.; Bidart, G. N.; Adams, P. D.; Dueber, J. E.; Svensson, B.; Rovira, C.; Welner, D. H. O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering. ACS Catal. 2021, 11 (11), 1810– 1815, https://doi.org/10.1021/acscatal.0c04171]
- Teze D, Bidart GN, and Welner DH. (2022). Family 1 glycosyltransferases (GT1, UGTs) are subject to dilution-induced inactivation and low chemo stability toward their own acceptor substrates. Front Mol Biosci. 2022;9:909659. DOI:10.3389/fmolb.2022.909659 |
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, and Davies GJ. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25(6):1396-405. DOI:10.1038/sj.emboj.7600970 |
- Mulichak AM, Losey HC, Walsh CT, and Garavito RM. (2001). Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9(7):547-57. DOI:10.1016/s0969-2126(01)00616-5 |
- Bourne Y and Henrissat B. (2001). Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol. 2001;11(5):593-600. DOI:10.1016/s0959-440x(00)00253-0 |
- Coutinho PM, Deleury E, Davies GJ, and Henrissat B. (2003). An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328(2):307-17. DOI:10.1016/s0022-2836(03)00307-3 |